April 26, 2006 Update: Version 1.21 Now Available. See the revision history for details.
(revision history)
- What is the FMRIB Plug-in for EEGLAB?
- What Methods Does the FMRIB plug-in Use?
- Requirements
- Download and Installation
- Tutorial
- Bugs and Suggestions
- Contributors
- Citing the FMRIB plug-in
- Licence
- References
What is the FMRIB Plug-in for EEGLAB?
The FMRIB plug-in is a set of Matlab tools developed at the University of Oxford Centre for Functional MRI of the Brain (FMRIB), which allow the removal of FMRI-related
artifacts from EEG data.
These tools are designed to work within the EEGLAB environment, providing
a GUI to remove FMRI gradient artifacts, detect QRS complexes from an ECG channel, and remove
pulse (ballistocardiographic/BCG) artifacts from the EEG. All of the tools
can also be used from the Matlab command line, providing expert users with the
ability to use them in custom scripts.
What Methods Does the FMRIB plug-in Use?
The methods used are based on published work in this area (see References) and more than two years of my own
(Rami Niazy) research in the field of
EEG/FMRI simultaneous experimentations. I am in the process of publishing a paper detailing methods and validations. For the mean time, the following section provides a very short summary of the methods.
FASTR: FMRI Artifact Slice Template Removal
FASTR [1] is the tool used to remove gradient artifacts from EEG data. It requires that a slice timing
event (or a section/volume trigger - beta) is present in the data i.e. an event for each FMRI slice acquired (for more
information about events please consult the EEGLAB website). The sampling rate of the data should be sufficient as to not
have any aliasing from high frequency gradient noise. All of our data were collected at 2048Hz (maximum gradient artifact frequency in a typical EPI
sequence is about 700-800Hz).
The first stage in FASTR is to align all the slice artifacts to correct for any slight jitter in the exact location of the slice-timing events. This is done by interpolating the data then shifting each artifact
until the correlation between it and a reference artifact (the first artifact in
the data) is maximised. This operation is done on the first EEG channel and the adjustments are
then applied to all the channels. The second stage is similar to the method
proposed by Allen et al. [2], in that an average template is computed for the artifact then subtracted. FASTR does this by taking a moving-window average of slice artifacts, then subtract the average template
from the centre contaminated data. For example, let us say EEG was
recorded during the acquisition of V FMRI volumes with S slices
per volume (a total of VxS =M slice artifacts). For a
contaminated EEG channel E, the data is divided into M segments
each with 1 slice artifact. For any segment k=1,2,3...M, we apply a
window that covers N+1 artifacts from Ek-N/2 to
Ek+N/2. The mean of the N+1 artifacts is then
computed to form the artifact template Ak. We can then
compute Ck=Ek-αAk, where Ck
is the cleaned EEG segment and α is a scalar that minimises the
difference (in a least squares sense) between Ck and Ek. Following this process, the
data is again divided according to the slice events and aligned in a matrix to calculate the principal components of the
artifact residuals using PCA. The first N (default 4) PCs are then taken to
form an optimal basis set (OBS) describing the variation in the artifact
residuals. The OBS is then fitted and subtracted from each segment.
Using OBS reduces any residual artifacts by up to 90%. The data are then low-pass filtered. As suggested by Allen et al. [2], adaptive noise cancellation
(ANC) is used in the final step to remove any remaining residuals. However, instead of using a binary
vector of slice timings (0s and 1s indicating slice timings) as a reference in the filter, FASTR uses the subtracted noise as a reference.
Using the subtracted noise as a reference in the ANC reduces the chances of removing useful information in the process.
QRS / Heart-Beat Detection
ECG collected in the MRI magnet usually suffers from augmented T segments due to the conductivity of blood.
This makes the identification of QRS events with simple thresholding
impractical. The identification of these QRS complexes locations is crucial to remove pulse/BCG artifacts. The FMRIB
plug-in provides a tool to robustly detect heart beats. The program uses combined adaptive thresholding [3] and the Teager energy operator [4], followed by a correction algorithm. This algorithm aligns all events and corrects for false positives and negatives.
The QRS / heart-beat detection tool has an average sensitivity and specificity
of 99%.
Pulse / BCG Artifact Removal
The removal of pulse/BCG artifacts uses QRS/Heart-beat events to subtract an artifact template from the data. Four methods are available for the formation of the artifact template. The first method is based on the same concept as the one used to remove the gradient residuals (OBS - see above). The user can also select to use the mean artifact [5], a gaussian-weighted mean artifact (to emphasise the shape of artifact being subtracted), or use the median artifact. Our results suggest that using OBS reduces the power of the residual pulse artifacts by an average of 50% over the other methods.
Those familiar with the ICA
functionality in EEGLAB, might be tempted to use ICA to remove pulse/BCG
artifacts in a similar manner to eye blink artifacts for example. However,
the nature of the pulse/BCG artifacts are such that the sources causing the
artifacts are not common to all electrodes and are moving/rotating sources,
which will cause ICA to fail as it is meant to deal with fixed, stationary
sources. To illustrate the moving/rotating sources idea, the
following movie shows the average topographical distribution of BCG artifacts
during a single heart beat:
 click on the thumbnail to play the movie.
click on the thumbnail to play the movie.
Movie is courtesy of Stefan Debener.
In addition to the requirements of EEGLAB, the FMRIB plug-in needs the Matlab Signal Processing Toolbox to work. The plug-in also requires an event indicating FMRI slice/segment timings for the removal of gradient artifacts i.e. an event type that occurs everytime an FMRI slice/volume is acquired. An ECG channel or an event indicating QRS complex / heart-beat occurrences is required for the removal of ballistocardiographic artifacts.
1) Download the compressed FMRIB plug-in file [tgz,
zip] into your 'plugins' directory of your EEGLAB
distribution.
2) Uncompress the downloaded file using utility software (gunzip,
tar).
Under UNIX: tar zxvf fmrib1.21.tgzInside your 'plugins' directory, you should now have a directory called 'fmrib1.21' containing all the necessary m-files and binaries. Starting EEGLAB should now automatically recognise and add the plug-in.
or unzip fmrib1.21.zip
Under WINDOWS: winzip
IMPORTANT NOTE REGARDING THE USE OF MEX-FILE BINARIES: The FMRIB plug-in uses matlab MEX files (Matlab callable c programs) to increase the processing speed. These files need to be compiled into binaries before they can be used. If a binary is not available for your machine architecture, the m-file version will be executed instead and things will still work properly, but slower. The download includes compiled binaries for Window, Linux, Linux x86_64 (complied on an Intel Xeon machine), and alpha 64-bit machines. The two programs that use MEX files are prcorr2.X and fastranc.X, which perform correlation and adaptive noise cancellation, respectively. 'X' is the extension denoting the architecture. An extention 'm' means this is the platform independent m-file, 'dll' is the windows binary, 'mexaxp' is the extention for alpha-64 processors, 'mexglx' is the linux binary, and 'mexa64' is the binary for 64-bit linux machines (although compiled on a Xeon, it should work on AMD 64 bit Operaton machines). To be absolutely sure of having the correct binary for your system, you can compile the binaries in Matlab if you have purchased/have a license for the Matlab compiler. If you do, run
>>mex -C prcorr2.c and
>>mex -C fastranc.c
from your MATLAB command line. Make sure you are in the 'fmrib1.21' directory. Otherwise, you will have to use with the much-slower M-file version of these two programs.
Tutorial
This short tutorial will take you through using the tools in this plug-in to process an example data set. I will assume you know the basics of EEGLAB e.g. loading a dataset, viewing, events, data structures,...etc. If not, refer to the EEGLAB website for more information.
1) First download the compressed example dataset (25MB) [tgz,
zip] and uncompress it as described in the Download and Installation section
above.
2) Start eeglab in MATLAB. You should see the line
eeglab: adding "fmrib1.21" plug-in (see >> help eegplugin_fmrib)
appear in your matlab environment window.
3) Load the example data set FMRIB_Data.set
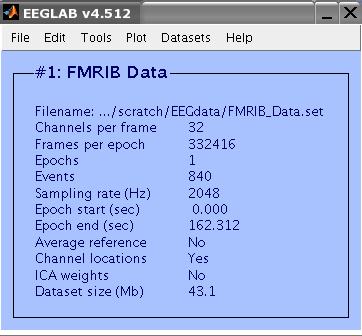
This a 32-Channel data collected during an FMRI recording session. The last two channels are EMG and ECG
channels. The data (including the EMG and ECG channels) were sampled at 2048 Hz. FMRI setting were TR=3s, 21
slices. The task performed by the subject was 10 seconds intervals of eyes opened/closed. Total FMRI experiment time was 2 minutes. There were 40 FMRI volumes collected, resulting in 840 slices. The data include an event type 'slice' for each slice collected.
4) View the data and scroll to the beginning of the gradient artifacts. You can see that the data is contaminated with gradient artifacts and line noise.
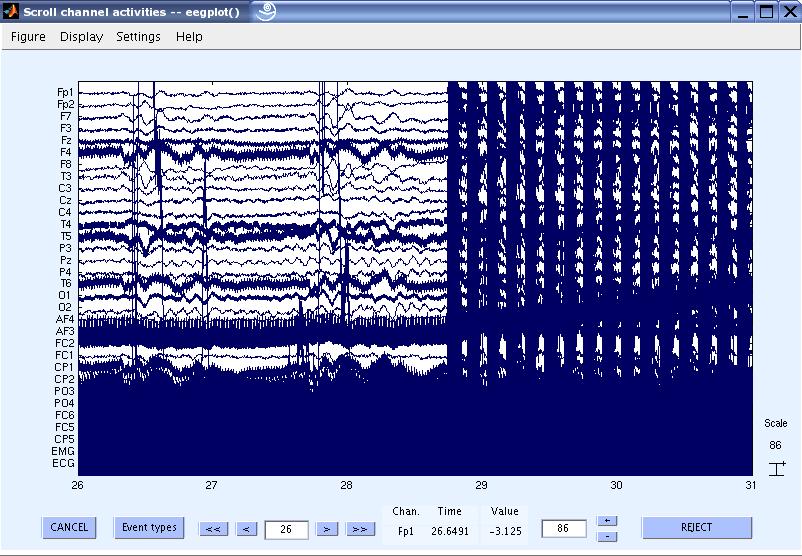
5) Now let us remove the gradient artifacts from the data. In the EEGLAB GUI, go to Tools
> FMRIB Tools > FASTR: Remove FMRI gradient artifacts.
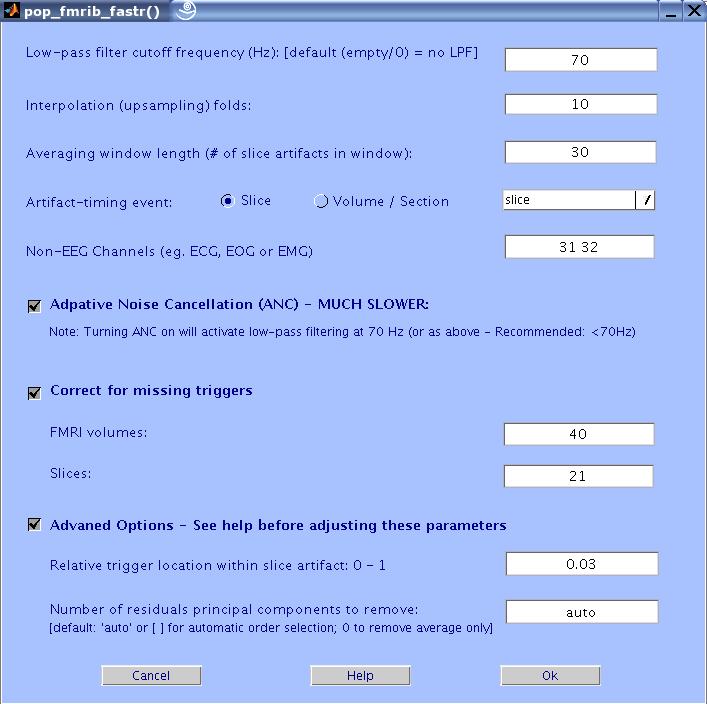
The FASTR GUI will now show up. Enter the values as in the figure below and press OK.
The Matlab window will now show the selected settings and show the progress of the process.
When FASTR is finished, you will be asked if you want to overwrite the parent
dataset or make a new one. Make a new dataset and call it 'FMRIB Data -
FASTR'.
Here is a description of the different GUI settings:
Low pass filter cutoff frequency (HZ): Low pass filtering is required to
remove high frequency gradient noise. Enter the desired cutoff frequency. Default: 70 Hz.
Interpolation (up-sampling) folds: In order to correctly remove the
gradient artifacts, the algorithm up-samples the data. It is
recommended that you up-sample the data to bring the sampling
frequency to about 20 kHz. The default up-sampling factor is 10.
Averaging window size: Enter how many realization of the slice artifacts are to be included in the averaging window. The bigger the
window the better the approximation of the true artifact is, but the
slower the adaptation is to any changes in the shape of the artifact
(due to head movement for example).
Artifact-timing event type: The program requires that the data in EEGLAB
have events that correspond to the timing of each FMRI artifact period (slice (optimal) or volume/section ) acquisition. The pop-up menu displays all available event types.
Select the one corresponding to the artifact timing and specify the type of triggers (slice or volume/section).
Adaptive Noise Cancellation (ANC): This checkbox turns on ANC. This
will make the entire process much slower. To use ANC, a low-pass
filter must be specified. If not, an LPF of 70Hz will be used.
Correct for missing triggers: Check this option if you want FASTR to add
missed triggers due to technical faults. The algorithm that corrects for
this needs to know the number of FMRI volumes and Slices collected
during the acquisition of the EEG data. It then calculates the
theoretical number of correct triggers to expect, and attempts to add any missing
ones. This feature has not been tested extensively, so use it with
caution. The drawback of not using it is that you could have
segments of artifact-contaminated data where the slice events are
missing.
Non-EEG Channels: Enter any channels in the data that are not EEG
channels. This is because fitting and removing the residual artefacts using PCA
and optimal basis set (OBS) might remove
features such as QRS complexes in ECG data.
The artifacts will still be removed but the residuals will not be further
processed
using OBS.
Advanced Options:
Relative trigger location: During acquisition of each FMRI slice, the
MRI machine sends a trigger. The time at which the trigger is sent
relative to the start of the slice acquisition may vary from one MRI
machine to another. In this interactive box, you can define the position
of the trigger relative to the start of the slice. A value of 0
indicates the trigger is at the exact beginning of the slice
collection and 1 indicates at the end. Usually the trigger is much
closer to the beginning than the end. The default value is 0.03.
Number of residual artifacts principal components to remove: FASTR (by default)
automatically estimates the number of components to remove. This is done by analysing the plot of ordered eigenvalues of the covariance matrix of the artifact residuals. The number of components is then (conservatively) selected for each channel depending on how much of the total variance is explained, how much of the variance each component explains, and the change of the variance explained from one component to another (the slope of the plot). If one chooses to fix the number of components to remove, it has been found that the first four PCs (at 2048 sampling) account for most of the residual
artifacts variance. If as few as 1 more PC is removed some EEG data will be
removed as well. It might be that for different datasets originally
sampled at different frequencies, different number of PCs is needed.
The best way to know is to do a PCA on a matrix of the slice
artifacts (after subtracting the artifact but before filtering) and
look at the eigenvalue plot. If you are not sure what that means or
how to do this use the default or experiment with different number of
PCs. Start with 2 and increase the number of PCs in steps of 1. TO SKIP THIS STEP, ENTER 0.
6) To save memory, we can now downsample the data to 256Hz. Go to Tools > Change sampling rate, and enter 256 as the new sampling rate. Click OK.
Store the date over the parent with the same name. NOTE: NOT DOWNSAMPLING TO A STANDARD EEG FREQUENCY (ABOUT 200 Hz OR SO) WILL CAUSE SUBSEQUENT STEPS TO BE VERY SLOW OR CRASH.
7) Now let us remove any line noise. Go to Tools > Filter the data, and enter [45 55] as the notch
filter range. Again, once done, store the data over the parent with the same name.
8) Now you can view the cleaned data. The red lines are the slice triggers indicating the start of the FMRI acquisition. You can also notice the high amplitude ECG signal. Scrolling through the data will also show the pulse / BCG artifacts correlated with the ECG.
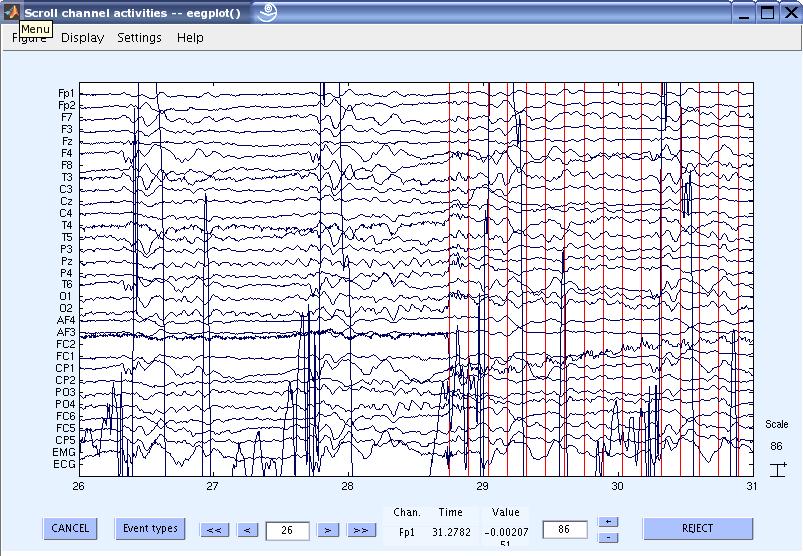
9) Now let us now remove the pulse / BCG artifact. First we need to detect the QRS complexes / Heart beats. Go to Tools
> FMRIB Tools > Detect QRS events.
Enter the parameters as in the figure below and click OK.
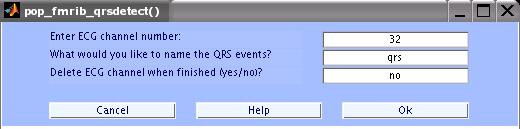
The MATLAB window will show the progress. When done, save the data as a new dataset with the name 'FMRIB Data - FASTR - QRS'.

10) The QRS events are now stored as type 'qrs'. We can now proceed with removing the pulse artifacts. Click on Tools
> FMRIB Tools > Remove pulse artifacts.
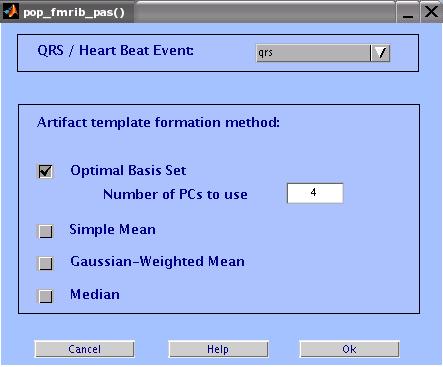
Enter the parameters as shown in the figure below, then click OK.
When the process is done, Store the data as a new dataset and call it 'FMRIB - CLEAN'.
Here is a description of the different available options in the GUI:
QRS / Heart Beat Events: This pop-up menu displays available event types
in the data. Select the event that corresponds to QRS complex locations
from ECG data.
Artifact Template Formation Method:
Optimal Basis Set: Does a PCA on a matrix of all the pulse artifacts
in a channel, then take the first N PCs to form an optimal basis
set (OBS) describing the variations in the artifact. The OBS is then
fitted and subtracted from each artifact. The default number of components is 4.
Simple Mean: Simply averages successive pulse artifacts around a
contaminated data segment then subtracts the result from the data.
Gaussian-Weighted Mean: Averages artifacts after multiplying by
a Gaussian window weights to emphasise current the artifact shape
and reduce effect of further artifacts.
Median: Uses a median filter to compute the artifacts template.
IMPORTANT NOTE: Many times, the quality of the ECG before fMRI scanning is different than that during scanning. High amplitude variations between the beginning of the data and rest of the data might cause instability in the QRS detection algorithm. It is advised that any data in the beginning of the recording that contains noise or is very different (ECG) from the rest of the data be discarded before QRS detection.
11) Done. Now scroll through the data to see the cleaned data. The data still has low frequency drifts, which can be removed by
high pass filtering using standard EEGLAB menus. If you take a look at the data about 10s after the start of FMRI (at about 39.5s) you will notice the alpha rhythms start and then
disappear after another 10s. This corresponds to the opening and closing of the eyes in the experiment.
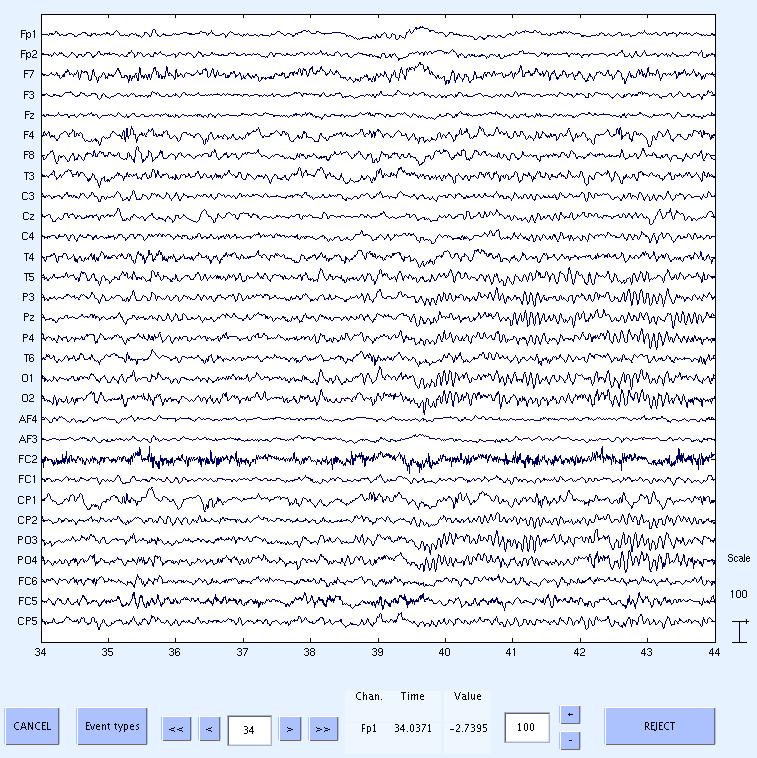
This concludes the tutorial.
Bugs and Suggestions
As I continue to update the plugin, I expect some bugs to show up. If this happens please send me a note. Also, please do not hesitate to contact me with suggestions or comments. I would also welcome any collaboration in extending the tools or adding new features.
If you have questions regarding the usage of the plug-in, please consult this
webpage and the 'help' documentation provided with the Matlab functions.
Contributors
This plug-in was written by Rami Niazy. Major technical advises were provided by
Christian Beckmann and Steve Smith. Testing and advises throughout were also
provided by Giandomenico Iannetti. Also, thanks to Stefan Debener for his continuous testing and evaluation of the plug-in and for providing suggestions, comments and
materials for the use in this webpage.
Valuable comments and suggestions about this webpage and the plug-in GUIs
were provided by Scott Makeig and Arnaud Delorme.
Citing the FMRIB plug-in
This is a free software distributed under the GPL. However, I do ask those that find this program of use to cite it in their work. A couple of publications about the tools used in this software
are in press. Please refer to it as the "FMRIB plug-in for EEGLAB, provided by the University of Oxford Centre for Functional MRI of the Brain (FMRIB)" and cite references [6,7]
below. Also, please make sure that EEGLAB is cited properly as described
in the EEGLAB website.
Licence
The FMRIB plug-in for EEGLAB, Release 1.21 (c) 2004-2006, The University of
Oxford (the "Software").
The Software remains the property of the University of Oxford ("the
University").
The FMRIB plug-in functions, sources and programs are released under the terms of the GPL
(http://www.gnu.org/copyleft/gpl.html).
The FMRIB plug-in is distributed "AS IS" under this Licence in the hope that it
will be useful, but in order that the University as a charitable
foundation protects its assets for the benefit of its educational and
research purposes, the University makes clear that no condition is
made or to be implied, nor is any warranty given or to be implied, as
to the accuracy of the Software, or that it will be suitable for any
particular purpose or for use under any specific conditions.
Furthermore, the University disclaims all responsibility for the use
which is made of the Software. It further disclaims any liability for
the outcomes arising from using the Software.
By downloading or making this software available to others you agree to the terms of this
licence and agree to let these terms known to other parties to whom you make this software available.
[1] Improved FMRI Artifact Reduction from Simultaneously
Acquired EEG Data using Slice Dependant Template Matching, Rami K. Niazy,
Giandomenico Iannetti, Christian Beckmann, Michael Brady, Stephen Smith.
Proc. 12th ISMRM (2004), Kyoto, Japan.
[2] A Method for Removing Imaging Artifact from Continuous EEG
Recording during Functional MRI, P.J. Allen, O. Josephs, R. Turner.
NeuroImage 12, 230-239 (2000).
[3] Real time electrocardiogram QRS detection using combined
adaptive threshold, Ivaylo I. Christov. Biomedical Engineering Online,
BioMed Central (2004). available at:
http://www.biomedical-engineering-online.com/content/3/1/28
[4] Improved ballistocardiac artifact removal from the
electroencephalogram recored in FMRI, KH Kim, HW Yoon, HW Park.
J Neourosience Methods 135 (2004) 193-203.
[5] Identification of EEG events in the MR
scanner: the problem of pulse artifact and a method for its
subtraction. Allen, P. et al. NeuroImage 8 229-239 (2002).
[6] Removal of FMRI environment artifacts from EEG data using optimal basis sets. R.K. Niazy et al. NeuroImage, in press (2005).
[7] Simultaneous recording of laser-evoked brain potentials and continuous, high-field functional magnetic resonance imaging in humans. G.D. Iannetti et al. NeuroImage, in press (2005).
This page was last updated September 16, 2005.
