Contents
- Introduction
- User Guide
Contents
- Before Running FEAT
- FEAT Basics
-
FEAT in Detail
- First-level or Higher-level Analysis?
- Full Analysis or Partial Analysis?
- Misc
- Data
- Pre-Stats
- Stats (First-level)
- Post-Stats: Contrasts, Thresholding, Rendering
- Registration
- Bottom Row of Buttons
- Time-Series Plots
-
Group Statistics
- Background and Theory
- FE and Within-Subject Multi-Session Analysis
- Setting Up Higher-Level Analysis in FEAT
- Single-Group Average (One-Sample T-Test)
- Unpaired Two-Group Difference (Two-Sample Unpaired T-Test)
- Randomise details
- Paired Two-Group Difference (Two-Sample Paired T-Test)
- Randomise details
- Tripled Two-Group Difference ("Tripled" T-Test)
- Multi-Session & Multi-Subject (Repeated Measures - Three Level Analysis)
- F-Tests
- Single-Group Average with Additional Covariate
- ANOVA: 1-factor 4-levels
- ANOVA: 1-factor 4-levels (Repeated Measures)
- ANOVA: 2-factors 2-levels
- ANOVA: 3-factors 2-levels
- Perfusion FMRI Analysis
- FEAT Output
- FEAT Programs
- Featquery - FEAT Results Interrogation
- Appendix A: Brief Overview of GLM Analysis
- Appendix B: Design Matrix Rules
- FAQ
Before Running FEAT
Before calling the FEAT GUI, you need to prepare each session's data as a 4D NIFTI or Analyze format image; there are utilities in fsl/bin called fslmerge and fslsplit to convert between multiple 3D images and a single 4D (3D+time) image. If the data requires any scanner-specific corrections (for example, for artefacts such as slice dropouts), this should be applied to the data before running FEAT.
Structural images for use as "highres" images in registration should normally be brain-extracted using BET before being used by FEAT.
FEAT Basics
To call the FEAT GUI, either type Feat in a terminal (type Feat_gui on Mac or Windows), or run fsl and press the FEAT button.
Now set the filename of the 4D input image (e.g. /users/sibelius/origfunc.nii.gz) by pressing Select 4D data. You can setup FEAT to process many input images, one after another, as long as they all require exactly the same analysis. Each one will generate its own FEAT directory, the name of which is based on the input data's filename (unless you enter an Output directory name).
Note that if you later run Statistics or if you are running Higher-level Analysis, then you can choose between selecting image files as the input or selecting FEAT directories. In the latter case first set the top two drop-down menus in the GUI and then select the FEAT directory or directories; it is important to select the FEAT directories before setting up anything else in FEAT. This is because quite a lot of FEAT settings are loaded from the first selected FEAT directory, possibly over-writing any settings which you might wish to change!
Total volumes (including volumes to be deleted) is automatically set from the input files chosen.
Now set Delete volumes. These should be the volumes that are not wanted because steady-state imaging is not reached for typically two or three volumes. These volumes are deleted as soon as FEAT is started, so any 4D data output by FEAT will not contain the deleted volumes. Note that Delete volumes should not be used to correct for the time lag between stimulation and the measured response - this is corrected for in the design matrix by convolving the input stimulation waveform with a blurring-and-delaying haemodynamic response function. Most importantly, remember when setting up the design matrix that the timings in the design matrix start at t=0 seconds, and this corresponds to the start of the first image taken after the deleted scans. In other words, the design matrix starts after the deleted scans have been deleted.
Set the TR (time from the start of one volume to the start of the next).
Now set the High pass filter cutoff point (seconds), that is, the longest temporal period that you will allow. A sensible setting in the case of an rArA or rArBrArB type block design is the (r+A) or (r+A+r+B) total cycle time. For event-related designs the rule is not so simple, but in general the cutoff can typically be reduced at least to 50s.
Note that most of the timing inputs in FEAT are set in seconds, not volumes. Total volumes and Delete volumes are exceptions.
Now click on the Stats tab and setup the model and required contrasts (for more detail see below).
When FEAT setup is complete and the Go button is pressed, the setup gets saved in a temporary FEAT setup file. Then a script (called feat - note the lower case) is run which uses the setup file and carries out all the FMRI analysis steps asked for, starting by creating a FEAT results directory, and copying the setup file into here, named design.fsf (this setup file can be later loaded back into FEAT using the Load button).
Once the script has started running you can Exit the FEAT GUI. The analysis will continue until completion, by default showing information about its progress in a web browser.
FEAT in Detail
First-level or Higher-level Analysis?
Use First-level analysis for analysing each session's data - i.e. the time-series analysis of the raw 4D FMRI data.
Use Higher-level analysis for combining first-level analyses. You can use this hierarchically - for example at second-level to analyse across several sessions and then at third-level to analyse across several subjects.
Full Analysis or Partial Analysis?
You can run a full analysis - Preprocessing and Statistics - or the individual stages separately
If you select Statistics, you may choose to select a a FEAT directory (or directories) instead of starting with 4D image data; the results already produced in those FEAT directories will then be used as appropriate.
Note that if you want to run only Statistics, you must select the FEAT directory/directories before editing the contrasts or thresholding parameters, as these will get reset on selection of the FEAT directory/directories.
Misc
Balloon help (the popup help messages in the FEAT GUI) can be turned off once you are familiar with FEAT.
The Progress watcher button allows you to tell Feat not to start a web browser to watch the FEAT analysis progress. If you are running lots of analyses you probably want to turn this off; you can view the same logging information by looking at the report_log.html files in any FEAT directories instead.
For the above two settings, you can control the default behaviour of the FEAT GUI by putting the following, with appropriate values set, in a file called .fslconf/feat.tcl in your home directory;
set fmri(help_yn) 1
set fmri(featwatcher_yn) 1
Brain/background threshold, % This is automatically calculated, as a % of the maximum input image intensity. It is used in intensity normalisation, brain mask generation and various other places in the FEAT analysis.
The next section controls the Design efficiency calculations made when setting up the EVs and contrasts in the paradigm design (see later for more on this). The Noise level % and Temporal smoothness together characterise the noise in the data, to be used only in the design efficiency estimation. The Noise level % is the standard deviation (over time) for a typical voxel, expressed as a percentage of the baseline signal level. The Temporal smoothness is the smoothness coefficient in a simple AR(1) autocorrelation model (much simpler than that actually used in the FILM timeseries analysis but good enough for the efficiency calculation here). If you want to get a rough estimate of this noise level and temporal smoothness from your actual input data, press the Estimate from data button (after you have told FEAT where your input data is). This takes about 30-60 seconds to estimate. This applies just the spatial and temporal filtering (i.e., no motion correction) that you have specified in the Pre-stats section, and gives a reasonable approximation of the noise characteristics that will remain in the fully preprocessed data, once FEAT has run. The Z threshold is the Z value used to determine what level of activation would be statistically significant, to be used only in the design efficiency calculation. Increasing this will result in higher estimates of required effect.
Cleanup first-level standard-space images; when you run a higher-level analysis, the first thing that happens is that first-level images are transformed into standard-space (in <firstlevel>.feat/reg_standard subdirectories) for feeding into the higher-level analysis. This takes up quite a lot of disk space, so if you want to save disk space, turn this option on and these these upsampled images will get deleted once they have been fed into the higher-level analysis. However, generating them can take quite a lot of time, so if you want to run several higher-level analyses, all using the same first-level FEAT directories, then leave this option turned off.
Data
First set the Number of inputs. At first level this is the number of identical analyses you want to carry out. At higher level this is the number of FEAT directories to be input from the lower-level analysis to the higher.
Next set the filename of the 4D input image (e.g. /home/sibelius/func.hdr). You can setup FEAT to process many input images, one after another, as long as they all require exactly the same analysis. Each one will generate its own FEAT directory, the name of which is based on the input data's filename.
Alternatively, if you are running either Statistics or running Higher-level analysis, the selection can be of either image files or FEAT directories. Note that in the latter case you should select the FEAT directories before setting up anything else in FEAT (such as changing the thresholds). This is because quite a lot of FEAT settings are loaded from the first selected FEAT directory, possibly over-writing any settings which you wish to change!
If the Output directory is left blank, the output FEAT directory name is derived from the input data name. (For higher-level analysis, the output name is derived from the first lower-level FEAT directory selected as input.) If, however, you wish to explicitly choose the output FEAT directory name, for example, so that you can include in the name a hint about the particular analysis that was carried out, you can set this here. This output directory naming behaviour is modified if you are setting up multiple first-level analyses, where you are selecting multiple input data sets and will end up with multiple output FEAT directories. In this case, whatever you enter here will be used and appended to what would have been the default output directory name if you had entered nothing. For example, if you are setting up 3 analyses with input data names /home/neo/fmri1.hdr, /home/neo/fmri2.hdr and /home/neo/fmri3.hdr, and set the output name to analysisA, the output directories will end up as /home/neo/fmri1_analysisA.feat etc.
Total volumes shows the number of FMRI volumes in the time series, including any initial volumes that you wish to delete. This will get set automatically once valid input data has been selected. Alternatively you can set this number by hand before selecting data so that you can setup and view a model without having any data, for experimental planning purposes etc.
Delete volumes controls the number of initial FMRI volumes to delete before any further processing. You should have decided on this number when the scans were acquired. Typically your experiment would have begun after these initial scans (sometimes called "dummy scans"). These should be the volumes that are not wanted because steady-state imaging has not yet been reached - typically two or three volumes. These volumes are deleted as soon as FEAT is started, so all 4D data sets produced by FEAT will not contain the deleted volumes. Note that Delete volumes should not be used to correct for the time lag between stimulation and the measured response - this is corrected for in the design matrix by convolving the input stimulation waveform with a blurring-and-delaying haemodynamic response function. Most importantly, remember when setting up the design matrix, that the timings in the design matrix start at t=0 seconds, and this corresponds to the start of the first image taken after the deleted scans. In other words, the design matrix starts AFTER the deleted scans have been deleted.
TR controls the time (in seconds) between scanning successive FMRI volumes.
The High pass filter cutoff controls the longest temporal period that you will allow. A sensible setting in the case of an rArA or rArBrArB type block design is the (r+A) or (r+A+r+B) total cycle time. For event-related designs the rule is not so simple, but in general the cutoff can typically be reduced at least to 50s. This value is setup here rather than in Pre-stats because it also affects the generation of the model; the same high pass filtering is applied to the model as to the data, to get the best possible match between the model and data.
Pre-Stats
You will normally want to apply Motion correction; this attempts to remove the effect of subject head motion during the experiment. MCFLIRT uses FLIRT (FMRIB's Linear Registration Tool) tuned to the problem of FMRI motion correction, applying rigid-body transformations. Note that there is no "spin history" (aka "correction for movement") option with MCFLIRT. This is because this is still a poorly understood correction method which is under further investigation.
B0 unwarping is now carried out using BBR, which also performs simultaneous registration to the structural image. Therefore this requires that registration and a structural (highres) image are specified. In this section you need to enter the B0 fieldmap images that should be created before you run FEAT and usually require site/scanner/sequence specific processing. See the FUGUE documentation for more information on creating these images. The fieldmap and functional (EPI) images must be in the same orientation (LR/AP/SI labels the same in FSLView), although they do not need to be registered or have the same resolution or exact FOV. In the GUI, the two images that are required are (1) a fieldmap image which must have units of rad/s, and (2) a brain-extracted and registered magnitude image (this is usually created by running BET on the standard magnitude-only reconstructed image from the fieldmap sequence data and should have all non-brain voxels removed to avoid noisy fieldmap values at the edge of the brain). FEAT will also look for the original non-brain-extracted fieldmap magnitude in the same directory as the brain-extracted image that you input into the GUI, this non-brain-extracted magnitude image should have the same filename as the brain-extracted version, with the only difference being the "_brain" at the end of the brain-extracted version. For example, fieldmap_mag and fieldmap_mag_brain, but not something like fieldmap_mag and fieldmap_mag_brain_v1, as the latter ends with "_brain_v1" and not just "_brain" and so this would cause a problem. Next you need to enter the Effective EPI echo spacing in milliseconds. This is the time between echoes in successive k-space lines. If you are using an accelerated sequence (parallel imaging) then the number you need here is the echo spacing for the acquired lines divided by the in-plane acceleration factor (note that multi-band accelerations, or similar, do not modify the effective echo spacing here unless combined with in-plane acceleration, in which case only the in-plane factor should be used). The EPI TE (echo time) is also in milliseconds. Both of these values relate to your FMRI EPI data, not the fieldmap data. You also need to specify the Unwarp direction, which is the phase-encoding direction of your FMRI EPI data (using voxel axes). The sign of this direction will depend on both the sign of the phase encode blips in the EPI sequence and on the sign of the fieldmap. As it can be difficult to predict this sign when using a particular site/scanner/sequence for the first time, it is usual to try both positive and negative values in turn and see which gives better undistortion (the wrong sign will increase the amount of distortion rather than decrease it). Finally, you need to specify a % Signal loss threshold. This determines where the signal loss in the EPI is too great for registration to get a good match between the EPI data and other images. Areas where the % signal loss in the EPI exceeds this threshold will get masked out of the registration process between the EPI and the fieldmap and structural images. If you are running both motion correction and B0 unwarping, the motion correction resampling does not get applied at the same time as the motion estimation; instead the motion correction gets applied simultaneously with the application of the B0 unwarping, in order to minimise interpolation-related image blurring. Once you have run FEAT you should definitely check the unwarping report (click on the mini-movie, shown in the main FEAT report page, that flicks between distorted and undistorted versions of example_func). In particular you should check that it looks like the unwarping has occurred in the correct direction (and change the unwarp direction and/or sign if it is not).
Slice timing correction corrects each voxel's time-series for the fact that later processing assumes that all slices were acquired exactly half-way through the relevant volume's acquisition time (TR), whereas in fact each slice is taken at slightly different times. Slice timing correction works by using (Hanning-windowed) sinc interpolation to shift each time-series by an appropriate fraction of a TR relative to the middle of the TR period. It is necessary to know in what order the slices were acquired and set the appropriate option here. If slices were acquired from the bottom of the brain to the top select Regular up. If slices were acquired from the top of the brain to the bottom select Regular down. If the slices were acquired with interleaved order (0, 2, 4 ... 1, 3, 5 ...) then choose the Interleaved option. If slices were not acquired in regular order you will need to use a slice order file or a slice timings file. If a slice order file is to be used, create a text file with a single number on each line, where the first line states which slice was acquired first, the second line states which slice was acquired second, etc. The first slice is numbered 1 not 0. If a slice timings file is to be used, put one value (ie for each slice) on each line of a text file. The units are in TRs, with 0 corresponding to no shift. Therefore a sensible range of values will be between -0.5 and 0.5.
By default BET brain extraction is applied to create a brain mask from the mean volume in the FMRI data. This is normally better than simple intensity-based thresholding for getting rid of unwanted voxels in FMRI data. Note that here, BET is setup to run in a quite liberal way so that there is very little danger of removing valid brain voxels. If the field-of-view of the image (in any direction) is less than 30mm then BET is turned off by default. Note that, with respect to any structural image(s) used in FEAT registration, you need to have already run BET on those before running FEAT.
Spatial smoothing is carried out on each volume of the FMRI data set separately. This is intended to reduce noise without reducing valid activation; this is successful as long as the underlying activation area is larger than the extent of the smoothing. Thus if you are looking for very small activation areas then you should maybe reduce smoothing from the default of 5mm, and if you are looking for larger areas, you can increase it, maybe to 10 or even 15mm. To turn off spatial smoothing simply set FWHM to 0. Note that the smoothing is performed via SUSAN, effectively providing Gaussian smoothing within the brain while excluding background voxels.
Intensity normalisation forces every FMRI volume to have the same mean intensity. For each volume it calculates the mean intensity and then scales the intensity across the whole volume so that the global mean becomes a preset constant. This step is normally discouraged - hence is turned off by default. When this step is not carried out, the whole 4D data set is still normalised by a single scaling factor ("grand mean scaling") - each volume is scaled by the same amount. This is so that higher-level analyses are valid.
Perfusion subtraction is a pre-processing step for perfusion FMRI (as opposed to normal BOLD FMRI) data. It subtracts even from odd timepoints in order to convert tag-control alternating timepoints into a perfusion-only signal. If you are setting up a full perfusion model (where you model the full alternating tag/control timeseries in the design matrix) then you should not use this option. The subtraction results in a temporal shift of the sampled signal to half a TR earlier; hence you should ideally shift your model forwards in time by half a TR, for example by reducing custom timings by half a TR or by increasing the model shape phase by half a TR. When you select this option, FILM prewhitening is turned off (because it is not well-matched to the autocorrelation resulting from the subtraction filter) and instead the varcope and degrees-of-freedom are corrected after running FILM in OLS mode. See the Perfusion section of the manual for more information.
Highpass temporal filtering uses a local fit of a straight line (Gaussian-weighted within the line to give a smooth response) to remove low frequency artefacts. This is preferable to sharp rolloff FIR-based filtering as it does not introduce autocorrelations into the data. Lowpass temporal filtering reduces high frequency noise by Gaussian smoothing (sigma=2.8s), but also reduces the strength of the signal of interest, particularly for single-event experiments. It is not generally considered to be helpful, so is turned off by default. By default, the temporal filtering that is applied to the data will also be applied to the model.
The MELODIC option runs the ICA (Independent Component Analysis) tool in FSL. We recommend that you run this, in order to gain insight into unexpected artefacts or activation in your data.
As well as being a good way to find structured noise (or unexpected activation) in your data, ICA can also be used to remove chosen components (normally obvious scanner-related or physiological artefacts) from your data in order, for example, in order to improve the FEAT results. In order to do this:
Run Feat pre-stats processing only, by selecting Pre-stats from the top-right menu. Make sure you turn on MELODIC ICA in the Pre-stats section of the FEAT GUI.
- Open the MELODIC report (feat_output_directory.feat/filtered_func_data.ica/report/00index.html) in a web browser and look through the components to identify those that you wish to remove; record the list of component numbers to remove.
- In a terminal, run the MELODIC denoising, using the commands:
cd feat_output_directory.feat
fsl_regfilt -i filtered_func_data -o denoised_data -d filtered_func_data.ica/melodic_mix -f "2,5,9"
where you should replace the comma-separated list of component numbers with the list that you previously recorded when viewing the MELODIC report.
Now reopen the FEAT GUI and set the top-right menu to Stats + Post-stats. Set the input data to be feat_output_directory.feat/denoised_data By default the final FEAT output directory will be in denoised_data.feat inside the first FEAT output directory.
Stats (First-level)
FILM General Linear Model
General linear modelling allows you to describe one or more stimulation types, and, for each voxel, a linear combination of the modelled response to these stimulation types is found which minimises the unmodelled noise in the fitting. If you are not familiar with the concepts of the GLM and contrasts of parameter estimates, then you should now read Appendix A.
For normal first-level time series analysis you should Use FILM prewhitening to make the statistics valid and maximally efficient. For other data - for example, very long TR (>30s) FMRI data, PET data or data with very few time points (<50) - this should be turned off.
You may want to include the head motion parameters (as estimated by MCFLIRT motion correction in the Pre-stats processing) as confound EVs in your model. This can sometimes help remove the residual effects of motion that are still left in the data even after motion correction. This is not strongly recommended as there is still much to learn about residual motion effects; simply adding such confound EVs is quite a simplistic solution. We would recommend instead turning on MELODIC in the FEAT Pre-stats and using ICA-based denoising as a better alternative to removing residual motion effects (see the FEAT manual for more information on that). However, if you do wish to include motion parameters in your model then select this option. If you do this, then once the motion correction has been run, the translation and rotation parameters are added as extra confound EVs in the model. If you select this option then only the components of the main EVs that are orthogonal to the motion confound EVs will be used in determining significance of the effects of interest. You cannot use this option unless you are carrying out both pre-stats and stats in the same FEAT run.
If you want to add other confound EVs than motion parameters, that you have precomputed for your data, then turn the Add additional confound EVs option on and then enter the filename of a raw text file (or, if you are setting up multiple analyses, enter one text file for each input FMRI dataset to be processed). The file can contain as many confound EVs as you like, each in a separate column in the text file. This option can be used to enter other forms of motion-related regressors, such as ones to remove outliers as created by the script fsl_motion_outliers.
Note that motion confound EVs and added additional confound EVs will have high pass and/or low pass temporal filtering applied before entry into the model, if this option is selected on the Pre-Stats tab.
There is also the (BETA) option to add voxelwise EVs as confounds (via a file containing a list of images representing individual voxelwise regressors). This option is designed to work naturally with the Physiological Noise Model (PNM) that creates EVs to model the effects of physiological noise (e.g. due to cardiac and respiratory processes). These EVs are different for each slice (voxel) due to the significant interaction between the slice-timing of the acquisition and these relatively fast physiological processes.
You can setup FILM easily for simple designs by pressing the Model setup wizard button. At first level, the options are regular rest-A-rest-A... or rest-A-rest-B-rest-A-rest-B... designs (block or single-event) for normal BOLD FMRI, or a rest-A-rest-A... design for full modelling of perfusion FMRI data. At second level, the options are one-group t-test, two-group-unpaired and two-group-paired t-tests. Then, in the wizard popup window, choose whether to setup rArA... or rArBrArB... designs (regular block or single-event). The r blocks will normally be rest (or control) conditions. The perfusion rArA... option sets up the full model for a simple perfusion experiment, setting up a constant-height control-tag EV, an average BOLD component EV and the interaction EV, which represents the control-tag functional modulation (see the Perfusion section of the manual for more information on this). Next, press Process and the model will be setup for you. If you want to setup a more complex model, or adjust the setup created by the wizard, press Full model setup button. This is now described in detail.
EVs
First set the Number of original EVs (explanatory variables) - basic number of explanatory variables in the design matrix; this means the number of different effects that you wish to model - one for each modelled stimulation type, and one for each modelled confound. For first-level analyses, it is common for the final design matrix to have a greater number of real EVs than this original number; for example, when using basis functions, each original EV gives rise to several real EVs.
Now you need to setup each EV separately. Choose the basic shape of the waveform that describes the stimulus or confound that you wish to model. The basic waveform should be exactly in time with the applied stimulation, i.e., not lagged at all. This is because the measured (time-series) response will be delayed with respect to the stimulation, and this delay is modelled in the design matrix by convolution of the basic waveform with a suitable haemodynamic response function (see below).
If you need an EV to be ignored, choose Empty (all zeros). You are most likely to want to do this if you want the EVs to all have the same meaning for multiple runs, but in some runs one or more EVs contain no events of the relevant type. Note that in this case you will get a warning about the matrix being rank deficient.
For an on/off (or a regularly-spaced single-event) experiment choose a square wave. To model single-event experiments with this method, the On periods will probably be small - e.g., 1s or even less.
For sinusoidal modelling choose the Sinusoid option and select the number of Harmonics (or overtones) that you want to add to the fundamental frequency.
For a single-event experiment with irregular timing for the stimulations, a custom file can be used. With Custom (1 entry per volume), you specify a single value for each timepoint. The custom file should be a raw text file, and should be a list of numbers, separated by spaces or newlines, with one number for each volume (after subtracting the number of deleted images). These numbers can either all be 0s and 1s, or can take a range of values. The former case would be appropriate if the same stimulus was applied at varying time points; the latter would be appropriate, for example, if recorded subject responses are to be inserted as an effect to be modelled. Note that it may or may not be appropriate to convolve this particular waveform with an HRF - in the case of single-event, it is.
For even finer control over the input waveform, choose Custom (3 column format). In this case the custom file consists of triplets of numbers; you can have any number of triplets. Each triplet describes a short period of time and the value of the model during that time. The first number in each triplet is the onset (in seconds) of the period, the second number is the duration (in seconds) of the period, and the third number is the value of the input during that period. The same comments as above apply, about whether these numbers are 0s and 1s, or vary continuously. The start of the first non-deleted volume correpsonds to t=0.
It is also possible to specify voxelwise EVs, which are defined by 4D images that contain the values for the EV at each time point and in each voxel.
Note that whilst ALL columns are demeaned before model fitting, neither custom format will get rescaled - it is up to you to make sure that relative scaling between different EVs is sensible. If you double the scaling of values in an EV you will halve the resulting parameter estimate, which will change contrasts of this EV against others.
If you select Interaction then the current EV is modelled as an interaction between other EVs, and is normally used to create a third EV from two existing EVs, to model the nonlinear interaction between two different conditions (or for a Psycho-Physiological Interaction, or PPI, analysis). On the line of buttons marked Between EVs you select which other EVs to interact to form the new one. The selected EVs then get multiplied together to form the current EV. Normally they are multiplied after (temporarily) shifting their values so that the minimum of each EV is zero (Make zero = Min); however, if you change the Make zero: option, individual EVs will instead be zero-centred about the min and max values (Centre) or de-meaned (Mean). If all EVs feeding into an interaction have the same convolution settings, the interaction is calculated before convolutions, and the same convolution applied to the interaction; if they do not all have the same settings, then all convolutions are applied before forming the interaction, and no further convolution is applied to the interaction EV.
For PPI analyses, you should probably do something like: Set EV1 to your main effect of interest, set EV2 to your data-derived regressor (with convolution turned off for EV2), and set EV3 to be an Interaction. EV3 would then be an interaction between EV1 and EV2, with EV1's zeroing set to Centre and EV2's zeroing set to Mean. For more detail on PPI analyses in theory and in practice, see Jill O'Reilly's PPI page.
If you have chosen a Square or Sinusoid basic shape, you then need to specify what the timings of this shape are. Skip is the initial period of zeros (in seconds) before the waveform commences. Off is the duration (seconds) of the "Off" periods in the square wave. On is the duration (seconds) of the "On" periods in the square wave. Period is the period (seconds) of the Sinusoid waveform. Phase is the phase shift (seconds) of the waveform; by default, after the Skip period, the square wave starts with a full Off period and the Sinusoid starts by falling from zero. However, the wave can be brought forward in time according to the phase shift. Thus to start with half of a normal Off period, enter the Phase as half of the Off period. To start with a full On period, enter the same as the Off period. Stop after is the total duration (seconds) of the waveform, starting after the Skip period. "-1" means do not stop. After stopping a waveform, all remaining values in the model are set to zero.
Convolution sets the form of the HRF (haemodynamic response function) convolution that will be applied to the basic waveform. This blurs and delays the original waveform, in an attempt to match the difference between the input function (original waveform, i.e., stimulus waveform) and the output function (measured FMRI haemodynamic response). If the original waveform is already in an appropriate form, e.g., was sampled from the data itself, None should be selected. The next three options are all somewhat similar blurring and delaying functions. Gaussian is simply a Gaussian kernel, whose width and lag can be altered. Gamma is a Gamma variate (in fact a normalisation of the probability density function of the Gamma function); again, width and lag can be altered. Double-Gamma HRF is a preset function which is a mixture of two Gamma functions - a standard positive function at normal lag, and a small, delayed, inverted Gamma, which attempts to model the late undershoot.
The remaining convolution options setup different basis functions. This means that the original EV waveform will get convolved by a "basis set" of related but different convolution kernels. By default, an original EV will generate a set of real EVs, one for each basis function.
The Optimal/custom option allows you to use a customised set of basis functions, setup in a plain text file with one column for each basis function, sampled at the temporal resolution of 0.05s. The main point of this option is to allow the use of "FLOBS" (FMRIB's Linear Optimal Basis Set), which is a method for generating a set of basis functions that has optimal efficiency in covering the range of likely HRF shapes actually found in your data. You can either use the default FLOBS set, or use the Make_flobs GUI on the FEAT Utils menu to create your own customised set of FLOBS.
The other basis function options, which will not in general be as good at fitting the data as FLOBS, are a set of Gamma variates of different widths and lags, a set of Sine waves of differing frequencies or a set of FIR (finite-impulse-response) filters (with FIR the convolution kernel is represented as a set of discrete fixed-width "impulses").
For all basis function options there is the option to force exact orthogonalisation of the functions with respect to each other. For basis functions which are generally expected to be orthogonal (normally just the Optimal/custom option) this option should normally be left on, otherwise you would normally expect to leave it turned off.
You should normally apply the same temporal filtering to the model as you have applied to the data, as the model is designed to look like the data before temporal filtering was applied. In this way, long-time-scale components in the model will be dealt with correctly. This is set with the Apply temporal filtering option.
Adding a fraction of the temporal derivative of the blurred original waveform is equivalent to shifting the waveform slightly in time, in order to achieve a slightly better fit to the data. Thus adding in the temporal derivative of a waveform into the design matrix allows a better fit for the whole model, reducing unexplained noise, and increasing resulting statistical significances. Thus, setting Add temporal derivative produces a new waveform in the final design matrix (next to the waveform from which it was derived) This option is not available if you are using basis functions.
Orthogonalising an EV with respect to other EVs means that it is completely independent of the other EVs, i.e. contains no component related to them. Most sensible designs are already in this form - all EVs are at least close to being orthogonal to all others. However, this may not be the case; you can use this facility to force an EV to be orthogonal to some or all other EVs. This is achieved by subtracting from the EV that part which is related to the other EVs selected here. In general the use of orthogonalisation is not recommended. However, one example useage is for convenient demeaning - if you had another EV which was a constant height spike train, and the current EV is derived from this other one, but with a linear increase in spike height imposed, to model an increase in response during the experiment for any reason. You would not want the current EV to contain any component of the constant height EV, so you would orthogonalise the current EV wrt the other.
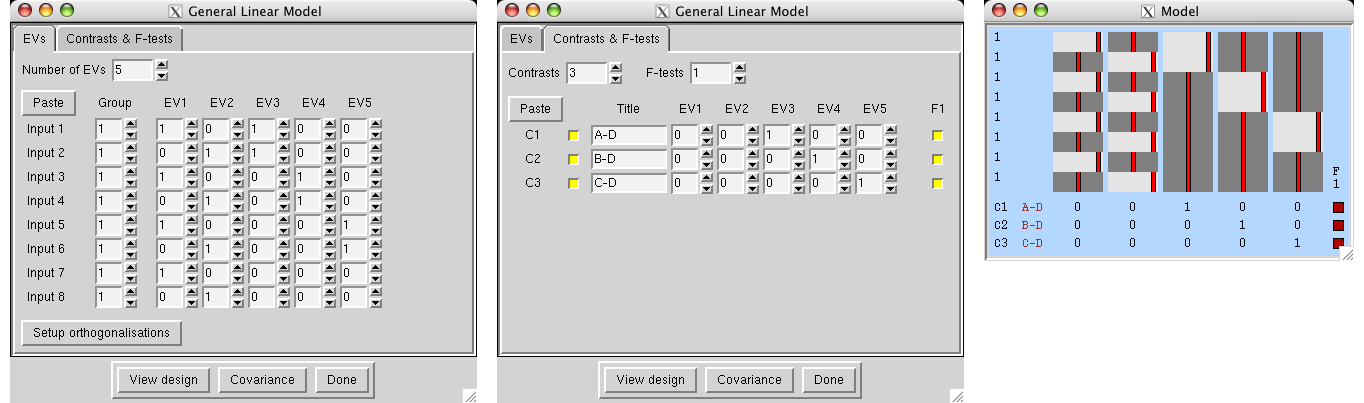
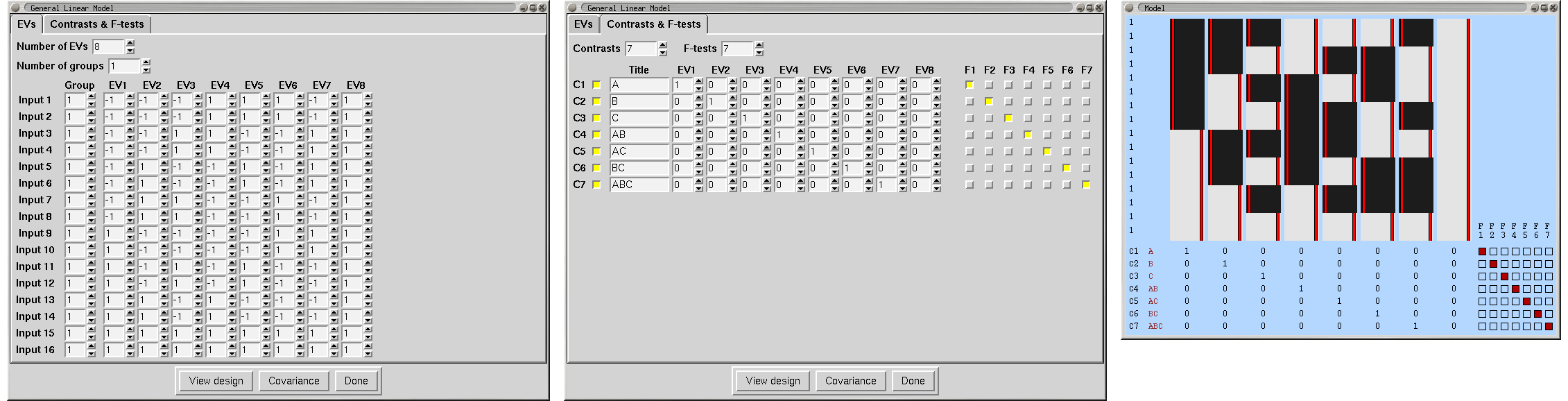
Contrasts
Each EV (explanatory variable, i.e., waveform) in the design matrix results in a PE (parameter estimate) image. This estimate tells you how strongly that waveform fits the data at each voxel - the higher it is, the better the fit. For an unblurred square wave input (which will be scaled in the model from -0.5 to 0.5), the PE image is equivalent to the "mean difference image". To convert from a PE to a t statistic image, the PE is divided by its standard error, which is derived from the residual noise after the complete model has been fit. The t image is then transformed into a Z statistic via standard statistical transformation. As well as Z images arising from single EVs, it is possible to combine different EVs (waveforms) - for example, to see where one has a bigger effect than another. To do this, one PE is subtracted from another, a combined standard error is calculated, and a new Z image is created.
All of the above is controlled by you, by setting up contrasts. Each output Z statistic image is generated by setting up a contrast vector; thus set the number of outputs that you want, using Number of contrasts. To convert a single EV into a Z statistic image, set its contrast value to 1 and all others to 0. Thus the simplest design, with one EV only, has just one contrast vector, and only one entry in this contrast vector; 1. To add more contrast vectors, increase the Number of contrasts. To compare two EVs, for example, to subtract one stimulus type (EV1) from another type (EV2), set EV1's contrast value to -1 and EV2's to 1. A Z statistic image will be generated according to this request.
For first-level analyses, it is common for the final design matrix to have a greater number of real EVs than the original number; for example, when using basis functions, each original EV gives rise to several real EVs. Therefore it is possible in many cases for you to setup contrasts and F-tests with respect to the original EVs, and FEAT will work out for you what these will be for the final design matrix. For example, a single [1] contrast on an original EV for which basis function HRF convolutions have been chosen will result in a single [1] contrast for each resulting real EV, and then an F-test across these. In general you can switch between setting up contrasts and F-tests with respect to Original EVs and Real EVs; though of course if you fine-tune the contrasts for real EVs and then revert to original EV setup some settings may be lost. When you View the design matrix or press Done at the end of setting up the model, an Original EVs setup will get converted to the appropriate Real EVs settings.
An important point to note is that you should not test for differences between different conditions (or at higher-level, between sessions) by looking for differences between their separate individual analyses. One could be just above threshold and the other just below, and their difference might not be significant. The correct way to tell whether two conditions or session's analyses are significantly different is to run a differential contrast like [1 -1] between them (or, at higher-level, run a higher-level FEAT analysis to contrast lower-level analyses); this contrast will then get properly thresholded to test for significance.
There is another important point to note when interpreting differential (eg [1 -1]) contrasts. This is that you are quite likely to only want to check for A>B if both are positive. Don't forget that if both A and B are negative then this contrast could still come out significantly positive! In this case, the thing to do is to use the Contrast masking feature (see below); setup contrasts for the individual EVs and then mask the differential contrast with these.
F-tests
F-tests enable you to investigate several contrasts at the same time, for example to see whether any of them (or any combination of them) is significantly non-zero. Also, the F-test allows you to compare the contribution of each contrast to the model and decide on significant and non-significant ones. F-tests are non-directional (i.e. test for "positive" and "negative" activation).
One example of F-test usage is if a particular stimulation is to be represented by several EVs, each with the same input function (e.g. square wave or custom timing) but all with different HRF convolutions - i.e. several basis functions. Putting all relevant resulting parameter estimates together into an F-test allows the complete fit to be tested against zero without having to specify the relative weights of the basis functions (as one would need to do with a single contrast). So - if you had three basis functions (EVs 1,2 and 3) the wrong way of combining them is a single (T-test) contrast of [1 1 1]. The right way is to make three contrasts [1 0 0] [0 1 0] and [0 0 1] and enter all three contrasts into an F-test. As described above, FEAT will automatically do this for you if you set up contrasts for original EVs instead of real EVs.
You can carry out as many F-tests as you like. Each test includes the particular contrasts that you specify by clicking on the appropriate buttons.
Buttons
To view the current state of the design matrix, press View design. This is a graphical representation of the design matrix and parameter contrasts. The bar on the left is a representation of time, which starts at the top and points downwards. The white marks show the position of every 10th volume in time. The red bar shows the period of the longest temporal cycle which was passed by the highpass filtering. The main top part shows the design matrix; time is represented on the vertical axis and each column is a different (real) explanatory variable (e.g., stimulus type). Both the red lines and the black-white images represent the same thing - the variation of the waveform in time. Below this is shown the requested contrasts; each row is a different contrast vector and each column refers to the weighting of the relevant explanatory variable. Thus each row will result in a Z statistic image. If F-tests have been specified, these appear to the right of the contrasts; each column is a different F-test, with the inclusion of particular contrasts depicted by filled squares instead of empty ones.
If you press Efficiency you will see a graphical representation of the covariance of the design matrix and the efficiency of the design/contrasts. Of most practical importance are the values in the lower part of the window, showing the estimability of the contrasts.
The first matrix shows the absolute value of the normalised correlation of each EV with each EV. If a design is well-conditioned (i.e. not approaching rank deficiency) then the diagonal elements should be white and all others darker. So - if there are any very bright elements off the diagonal, you can immediately tell which EVs are too similar to each other - for example, if element [1,3] (and [3,1]) is bright then columns 1 and 3 in the design matrix are possibly too similar. Note that this includes all real EVs, including any added temporal derivatives, basis functions, etc. The second matrix shows a similar thing after the design matrix has been run through SVD (singular value decomposition). All non-diagonal elements will be zero and the diagonal elements are given by the eigenvalues of the SVD, so that a poorly-conditioned design is obvious if any of the diagonal elements are black.
In the lower part of the window, for each requested contrast, that contrast's efficiency/estimability is shown. This is formulated as the strength of the signal required in order to detect a statistically significant result for this contrast. For example, in FMRI data and with a single regressor, this shows the BOLD % signal change required. In the case of a differential contrast, it shows the required difference in BOLD signal between conditions. This Effect Required depends on the design matrix, the contrast values, the statistical significance level chosen, and the noise level in the data (see the Misc tab in the main FEAT GUI). The lower the effect required, the more easily estimable is a contrast, i.e. the more efficient is the design. Note that this does not tell you everything that there is to know about paradigm optimisation. For example, all things being equal, event-related designs tend to give a smaller BOLD effect than block designs - the efficiency estimation made here cannot take that kind of effect into account!
When you have finished setting up the design matrix, press Done. This will dismiss the GLM GUI, and will give you a final view of the design matrix.
Post-Stats: Contrasts, Thresholding, Rendering
If you choose a mask for Pre-threshold masking then all stats images will be masked by the chosen mask before thresholding. There are two reasons why you might want to do this. The first is that you might want to constrain your search for activation to a particular area. The second is that in doing so, you are reducing the number of voxels tested and therefore will make any multiple-comparison-correction in the thresholding less stringent. The mask image chosen does not have to be a binary mask - for example, it can be a thresholded stats image from a previous analysis (in the same space as the data to be analysed here); only voxels containing zero in the mask image will get zeroed in this masking process. If pre-threshold masking is used, it is still necessary to carry out thresholding.
Thresholding: After carrying out the initial statistical test, the resulting Z statistic image is then normally thresholded to show which voxels or clusters of voxels are activated at a particular significance level.
If Cluster thresholding is selected, a Z statistic threshold is used to define contiguous clusters. Then each cluster's estimated significance level (from GRF-theory) is compared with the cluster probability threshold. Significant clusters are then used to mask the original Z statistic image for later production of colour blobs. This method of thresholding is an alternative to Voxel-based correction, and is normally more sensitive to activation. You may well want to increase the cluster creation Z threshold if you have high levels of activation.
The FEAT web page report includes a table of cluster details, viewed by clicking on the relevant colour-overlay image. Note that cluster p-values are not given for contrasts where post-threshold contrast masking (see below) is applied, as there is not a sensible p-value associated with the new clusters formed after masking.
If Voxel thresholding is selected, GRF-theory-based maximum height thresholding is carried out, with thresholding at the level set, using one-tailed testing. This test is less overly-conservative than Bonferroni correction.
You can also choose to simply threshold the uncorrected Z statistic values, or apply no thresholding at all.
Contrast masking: You can setup the masking of contrasts by other contrasts; after thresholding of all contrasts has taken place you can further threshold a given Z statistic image by masking it with non-zeroed voxels from other contrasts.
This means that of the voxels which passed thresholding in the contrast (or F-test) of interest, only those which also survived thresholding in the other contrasts (or F-tests) are kept.
As a further option, the generated masks can be derived from all positive Z statistic voxels in the mask contrasts rather than all voxels that survived thresholding.
Rendering: The Z statistic range selected for rendering is automatically calculated by default, to run from red (minimum Z statistic after thresholding) to yellow (maximum Z statistic). If more than one colour rendered image is to be produced (i.e., when multiple constrasts are created) then the overall range of Z values is automatically found from all of the Z statistic images, for consistent Z statistic colour-coding.
If multiple analyses are to be carried out separately, Use preset Z min/max should be chosen, and the min/max values set by hand. Again, this ensures consistency of Z statistic colour-coding - if several experiments are to be reported side-by-side, colours will refer to the same Z statistic values in each picture. When using this option, you should choose a conservatively wide range for the min and max (e.g., min=1, max=15), to make sure that you do not carry out unintentional thresholding via colour rendering.
With Solid colours you don't see any sign of the background images within the colour blobs; with Transparent colours you will see through the colour blobs to the background intensity.
If you are running a Higher-level analysis you can select what image will be used as the background image for the activation colour overlays. The default of Mean highres is probably the best for relating activation to underlying structure. For a sharper underlying image, (but one which is not so representative of the group of subjects), you can instead choose to use the highres image from the first selected subject. You can alternatively choose to use the original lowres functional data for the overlays, or the standard-space template image.
Registration
Before any multi-session or multi-subject analyses can be carried out, the different sessions need to be registered to each other. This is made easy with FEAT, by saving the appropriate transformations inside the FEAT directories; the transformations are then applied when group statistics is carried out, to tranform any relevant statistic images into the common space. By doing this (saving the relevant registration transformations and only applying them to the stats images later) a lot of disk space is saved.
Registration inside FEAT uses FLIRT (FMRIB's Linear Image Registration Tool). This is a very robust affine registration program which can register similar type images (intra-modal) or different types (inter-modal). Optionally, the structural to standard space registration can then be further refined by using FNIRT(FMRIB's Nonlinear Image Registration Tool), a fast and accurate nonlinear registration program.
Typically, registration in FEAT is a two-stage process. First an example FMRI low resolution image is registered to an example high resolution image (normally the same subject's T1-weighted structural). The transformation for this is saved into the FEAT directory. Then the high res image is registered to a standard image (normally a T1-weighted image in standard space, such as the MNI 152 average image). This transformation, also, is saved. Finally, the two transformations are combined into a third, which will take the low resolution FMRI images (and the statistic images derived from the first-level analyses) straight into standard space, when applied later, during group analysis.
For the registration of EPI to the structural image the recommended method is to use the BBR cost function, ideally (but optionally) with fieldmaps (requiring appropriate inputs in the Pre-Stats tab). This has been found to give substantially better registrations.
The Expanded functional image is an image which the low resolution functional data will be registered to, and this in turn will be registered to the main highres image. It only makes sense to have this "expanded functional" if a main highres image is also specified and used in the registration. One example of an expanded highres structural image might be a medium-quality structural scan taken during a day's scanning, if a higher-quality image has been previously taken for the subject. A second example might be a full-brain EPI image with the same MR sequence as the functional data, useful if the actual functional data is only partial-brain. It is strongly recommended that this image have non-brain structures already removed, for example by using BET.
If the field-of-view of the functional data (in any direction) is less than 120mm, then the registration of the functional data will by default have a reduced degree-of-freedom, for registration stability.
If you are attempting to register partial field-of-view functional data to a whole-brain image then 3 DOF is recommended - in this case only translations are allowed.
If the orientation of any image is different from any other image it may be necessary to change the search to Full search.
The Main structural image is is the main high resolution structural image which the low resolution functional data will be registered to (optionally via the initial structural image), and this in turn will be registered to the standard brain. It is strongly recommended that this image have non-brain structures already removed, for example by using BET.
Standard space refers to the standard (reference) image; it should be an image already in standard space, ideally with the non-brain structures already removed.
If you turn on Nonlinear then FNIRT will be used to apply nonlinear registration between the subject's structural image and standard space. FLIRT will still be used before FNIRT, to initialise the nonlinear registration. Nonlinear registration only works well between structural images and standard space; you cannot use it without specifying a structural image. FNIRT requires whole head (non-brain-extracted) input and reference images for optimal accuracy; if you turn on nonlinear registration, FEAT will look for the original non-brain-extracted structural and standard space images in the same directory as the brain-extracted images that you input into the GUI, these non-brain extracted images should have the same filename as the brain-extracted version appended with \"_brain\" at the end. It will complain if it can't find these, and if this is not corrected, nonlinear registration will run using the brain-extracted images, which is suboptimal. The Warp resolution controls the degrees-of-freedom (amount of fine detail) in the nonlinear warp; it refers to the spacing between the warp field control points. By increasing this you will get a smoother ("less nonlinear") warp field and vice versa.
In the case that a registration does not work well (and you should always visually check the quality of your registrations) it may be necessary to customise the registration by running different types of registration options outside of FEAT. If this is done then these customised registrations can be incorporated back into the FEAT analysis by using the updatefeatreg script.
Bottom Row of Buttons
When you have finished setting up FEAT, press Go to run the analysis. Once FEAT is running, you can either Exit the GUI, or setup further analyses.
The Save and Load buttons enable you to save and load the complete FEAT setup to and from file. The filename should normally be chosen as design.fsf - this is also the name which FEAT uses to automatically save the setup inside a FEAT directory. Thus you can load in the setup that was used on old analyses by loading in this file from old FEAT directories.
The Utils button produces a menu of FEAT-related utilities:
Load FEAT results into MEDx (only seen if FEAT is run from within MEDx). This opens a new folder and loads in results from a FEAT directory, setting up each stats image to be "time series clickable".
Featquery is a program which allows you to interrogate FEAT results by defining a mask or set of co-ordinates (in standard-space, highres-space or loweres-space) and get mean stats values and time-series.
Simple stats colour rendering allows you to overlay one or two stats images on a background image of the same size.
Colour render FEAT stats in high res produces colour rendered stats images in a selected FEAT directory, using either the original high resolution structural image as the background, or the structural image transformed into standard space as the background. This script starts by transforming stats into high resolution space and then produces 3D colour overlay images. 2D pictures of these are then saved to file and the 3D files removed to save disk space, but this removal can be turned off in the GUI.
Time-Series Plots
FEAT generates a set of time-series plots for data vs model for peak Z voxels. The main FEAT report web page contains a single plot for each contrast (from the peak voxel); clicking on this takes you to more plots related to that contrast, including also, in the case of cluster-based thresholding, plots averaged over all significant voxels.
Plots of full model fit vs data show the original data and the complete model fit given by the GLM analysis.
Plots of cope partial model fit vs reduced data show the model fit due simply to the contrast of interest versus that part of the data which is relevant to the reduced model (i.e. full data minus full model plus cope partial model). This generally is only easily interpretable in the case of simple non-differential contrasts.
Peri-stimulus plots show the same plots as described above but averaged over all "repeats" of events, whether ON-OFF blocks in a block-design, or events in an event-related design. Thus you get to see the average response shape. Note that FEAT tries to guess what an "event" is in your design automatically, so in complex designs this can give somewhat strange looking plots! The peri-stimulus plots are for the peak voxel only; one pair of full/partial plots is produced for each EV in the design matrix for that peak voxel, with the "events" defined from that EV only.
Group Statistics
Background and Theory
For higher-level analysis (e.g. analysis across sessions or across subjects) FEAT uses FLAME (FMRIB's Local Analysis of Mixed Effects). FLAME uses sophisticated methods for modelling (see related techreport TR01CB1) and estimating the inter-session or inter-subject random-effects component of the mixed-effects variance, by using MCMC to get an accurate estimation of the true random-effects variance and degrees of freedom at each voxel (see related techreport TR03MW1).
FEAT offers both fixed effects (FE) and mixed effects (ME) higher-level modelling. FE modelling is more "sensitive" to activation than ME, but is restricted in the inferences that can be made from its results; because FE ignores cross-session/subject variance, reported activation is with respect to the group of sessions or subjects present, and not representative of the wider population. ME does model the session/subject variability, and it therefore allows inference to be made about the wider population from which the sessions/subjects were drawn.
The remainder of this section discusses FLAME's mixed effects modelling.
"Mixed-effects" (ME) variance is the sum of "fixed-effects" (FE) variance (the within-session across-time variances estimated in the first-level analyses) and "random-effects" (RE) variance (the "true" cross-session variances of first-level parameter estimates). Note that the labels "mixed effects" and "random effects" are often (incorrectly) used interchangeably, partly because they are in practice often (but, importantly, not always) quite similar.
One factor that makes FEAT's approach to higher-level modelling particularly powerful is that it is easy to model and estimate different variances for different groups in the model. For example, an unpaired two-group comparison (e.g. between controls and patients) can be analysed with separate estimates of variance for each group. It is simply a case of specifying in the GUI what group each subject belongs to. (Note - FLAME does not model different group variances differently in the case of higher-level F-tests, due to the complexity of the resulting distributions; this may be addressed in the future.)
A second sophistication not normally available in multi-level analyses is the carrying-up of the first-level (FE) variances to the higher-level analyses. This means that the FE component of the higher-level ME variance can be taken into account when attempting to estimate the ME variance. One reason why it is suboptimal to simply use the directly-estimated ME variance is that this is often in practice lower than the estimated FE variance, a logical impossibility which implies negative RE variance. FEAT forces the RE variance in effect to be non-negative, giving a better estimate of ME variance.
Another reason for wanting to carry up first-level variances to the higher-level analyses is that it is not then necessary for first-level design matrices to be identical (ie "balanced designs" - for example having the same number of time points or event timings). (Note though: the "height" of design matrix waveforms at first-level must still be compatible across analyses.)
A third advantage in higher-level analysis with FEAT is that it is not necessary for different groups to have the same number of subjects (another aspect to design balancing) for the statistics to be valid, because of the ability to model different variances in different groups.
The higher-level estimation method in FEAT (FLAME) uses the above modelling theory and estimates the higher-level parameter estimates and ME variance using sophisticated estimation techniques. First, the higher-level model is fit using a fast approximation to the final estimation ("FLAME stage 1"). Then, all voxels which are close to threshold (according to the selected contrasts and thresholds) are processed with a much more sophisticated estimation process involving implicit estimation of the ME variance, using MH MCMC (Metropolis-Hastings Markov Chain Monte Carlo sampling) to give the distribution for higher-level contrasts of parameter estimates, to which a general t-distribution is then fit. Hypothesis testing can then be carried out on the fitted t-distribution to give inference based upon the best implicit estimation of the ME variance.
FE and Within-Subject Multi-Session Analysis
One use of the fixed-effects option is for combining multi-session analyses within-subject. You may want to change the normal way of running registration at single-level, if you would rather do the FE analysis at the resolution of the subject's structural MR image, and overlay the multi-session FE activation on this. To do this, when running first-level registrations, turn off the Main structural image option and select the (brain-extracted) structural image as Standard space.
Note that if you also want to feed the first-level data into multi-subject analyses, you will then want to redo the (first-level) registrations in the normal way.
Setting Up Higher-Level Analysis in FEAT
For figures showing the file and directory structures for first- and second-level FEAT analyses, see the Output section.
First change First-level analysis to Higher-level analysis. Note that the only processing option available is Stats + Post-stats; this is because at higher-level both Stats and Post-stats always need setting up, as the thresholding to be carried out affects the functioning of the core stats estimation. For the same reason it is not possible to re-threshold a higher-level analysis - the whole higher-level analysis must be re-run.
You can choose whether your higher-level design matrix will be applied to a set of lower-level cope images or a set of lower-level FEAT directories. In the latter, more normal, case, each contrast in the lower-level FEAT directories will have the higher-level model applied, each resulting in its own FEAT directory within the new group FEAT directory.
Now set the Number of analyses and Select FEAT directories (the first-level FEAT analyses to be fed into this higher-level analysis). FEAT will produce a new directory containing the group stats results; unless you specify an output directory name, the output directory name will be derived from the name of the first selected first-level FEAT directory. The suffix .gfeat is used.
Now setup the Stats. The main choice here is between fixed effects (FE) and mixed effects (ME) higher-level modelling. FE modelling is more "sensitive" to activation than ME, but is restricted in the inferences that can be made from its results; because FE ignores cross-session/subject variance, reported activation is with respect to the group of sessions or subjects present, and not representative of the wider population. ME does model the session/subject variability, and it therefore allows inference to be made about the wider population from which the sessions/subjects were drawn.
The FE option implements a standard weighted fixed effects model. No random effects variances are modelled or estimated. The FE error variances are the variances (varcopes) from the previous level. Weighting is introduced by allowing these variances to be unequal (heteroscedastic). Degrees-of-freedom are calculated by summing the effective degrees-of-freedom for each input from the previous level and subtracting the number of higher-level regressors.
We now discuss the different ME options.
OLS (ordinary least squares) is a fast estimation technique which ignores all lower-level variance estimation and applies a very simple higher-level model. This is the least accurate of the ME options.
For the most accurate estimation of higher-level activation you should use FLAME (FMRIB's Local Analysis of Mixed Effects) modelling and estimation. This is a sophisticated two-stage process using Bayesian modelling and estimation (for example it allows separate modelling of the variance in different subject groups, and forces random effects variance to be non-negative).
The first stage of FLAME is significantly more accurate than OLS, and nearly as fast. The second stage of FLAME increases accuracy slightly over the first stage, but is quite a lot slower (typically 45-200 minutes). It takes all voxels which FLAME stage 1 shows to be near threshold and carries out a full MCMC-based analysis at these points, to get the most accurate estimate of activation.
We generally recommend using FLAME 1, as it is MUCH faster than running both stages, and nearly as accurate. The added value from running full FLAME 1+2 is most significant in a highest-level analysis when you have a small number of subjects (say <10).
If you are carrying out a mid-level analysis (e.g., cross-sessions) and will be feeding this into an even higher-level analysis (e.g., cross-subjects), then you should not use the FLAME 1+2 option, as it is not possible for FLAME to know in advance of the highest-level analysis what voxels will ultimately be near threshold. With respect the question of whether to use fixed-effects or mixed-effects for such mid-level analyses, it could be argued that a mixed-effects analysis should be done at the mid-level. A mixed-effects analysis would assume that the sessions are randomly sampled from a "population" of sessions that that subject could produce. This includes estimation of each subject's session-to-session variance. However, it is common for only a small number of sessions to be collected for each subject, making estimation of each subject's session-to-session variance impractical. One solution to this is to assume a common session-to-session variance for all subjects, thereby providing enough data for the session-to-session variance to be estimated. However, this has a downside in that you lose information about which subjects are good (i.e. low variance) and which subjects are bad (i.e. high variance). Hence, when only a small number of sessions has been collected for each subject (say, less than 10), it is recommended that you use a fixed-effects analysis at the mid-level. This in effect treats the multiple first-level sessions (for each subject) as if they were one long session. Although this does ignore the session-session variability, it is arguable that this is not of interest anyway (this is a somewhat philosophical debate). In short, fixed-effects is favoured as it avoids practical problems associated with esimating the session-to-session variance (when there are not many sessions per subject), at the same time as maintaining information about which subjects are good and bad.
If you do decide to run FLAME 1+2 and the FEAT logs indicate a large difference between the stage 1 and stage 2 estimations (or, for example, the final thresholded zstat image looks "speckled"), this is an indication that your data is highly non-Gaussian (e.g., has one or more strong outlier subjects, or has two clearly different groups of subjects being modelled as a single group). In such a case, stage 1 estimation is quite inaccurate (OLS even more so), hence the larger-than-normal difference between stages 1 and 2. The only really good solution is to investigate in your data what is going on - for example, to find the bad outlier.
If you turn on Use automatic outlier de-weighting then FLAME will automatically detect outlier datapoints (for each voxel, each subject's data is considered with respect to the other subjects regarding whether it appears to be an outlier) [Woolrich M (2008), NeuroImage 41(2)]. Outliers are then automatically de-weighted in the multi-subject statistics. Outlier de-weighting is only available for the mixed effects options as it doesn't make sense in the context of a fixed effects model. It inceases the computation time considerably. The estimated outlier behaviour is stored in the stats directory found inside the higher-level FEAT directory. The prob_outlier1.nii.gz file is a 4D file giving the probability that each subject has outlier data on a voxelwise basis. The global_prob_outlier1.nii.gz file is a 3D file that indicates the size of the outlier population expressed as the proportion of subjects that are outliers (see the paper for more details). Note there are versions of these files for each variance group in the analysis.
For certain higher-level design types you can use the "wizard" - press the Model setup wizard button. Then choose whether to setup single group average or two groups, unpaired or two groups, paired designs. In the case of the unpaired two-group test, set the number of subjects in the first group. When you then press Process the design will be setup for you.
Next go to the full model setup. First choose the Number of EVs (different effects to be modelled). Next, the Number of groups is the number of different groups (of lower-level sessions or subjects). If you ask for more than one group, each group will end up with a separate estimate of variance for the higher-level parameter estimates; for example, if the first 10 inputs are first-level FEAT outputs from control subjects and the next 10 inputs are first-level FEAT outputs from patients, you can setup two different groups and each will end up with its own variance estimates, possibly improving the final modelling and estimation quality (see examples below for further clarification). If you setup different groups for different variances, you will get fewer data-points to estimate each variance (than if only one variance was estimated). Therefore, you only want to use this option if you do believe that the groups possibly do have different variances. If you setup different groups for different variances, it is necessary that, for each EV, only one of the sub-groups has non-zero values. Thus, for example, in the case of an unpaired t-test:
GP EV1 EV2 1 1 1 1 1 1 1 1 1 1 1 1 2 1 -1 2 1 -1 2 1 -1
is wrong with respect to this issue, and the following is correct:
GP EV1 EV2 1 1 0 1 1 0 1 1 0 1 1 0 2 0 1 2 0 1 2 0 1
Unlike with first-level analyses, the data (and the model) does not get demeaned. This is because mean effects are usually of interest! One effect of this is that a two-group unpaired model needs 2 EVs - one for finding each group's mean; it will not work to have a single EV containing 1's and -1's.
Setting the Number of additional, voxel-dependent EVs allows you to add voxel-dependent EVs; every voxel will have a different higher-level model. For each additional EV that you ask for, you will need to provide the filename of a 4D image file whose first 3 dimensions are the size of standard space, and whose 4th dimension corresponds to the number of sessions/subjects that you are inputting into this higher-level analysis. A typical use of voxel-dependent EVs would be to insert grey-matter partial volume information on the basis of structural imaging. Note that when you use this option and view the design matrix, a voxel-dependent EV is respresented graphically by the mean EV across all voxels, which may well not be very meaningful. If you want to use structural images (as used in the first-level FEAT registrations) to create the covariates, then you can easily generate the 4D covariate image with the feat_gm_prepare script; just type the script name followed by the desired 4D output image name and then the full list of first-level FEAT directories (these must be in the same order as they will appear as inputs to the higher-level FEAT analysis). You should run this script after all the first-level FEAT analyses and before running the higher-level FEAT.
Now setup the required Contrasts & F-tests and Post-stats (see examples below).
The higher-level design matrix is applied separately to each of the lower-level contrasts; thus each lower-level contrast will result in a new FEAT directory within the new top-level group FEAT directory. When FEAT has completed the higher-level analyses the new top-level group FEAT output directory contains a web page report which contains: a link to each of the original lower-level FEAT directories; a link to each of the higher-level FEAT analyses (one for each lower-level contrast); the picture of the higher-level design matrix.
We now give specific examples of how to set up the most common high-level analyses.
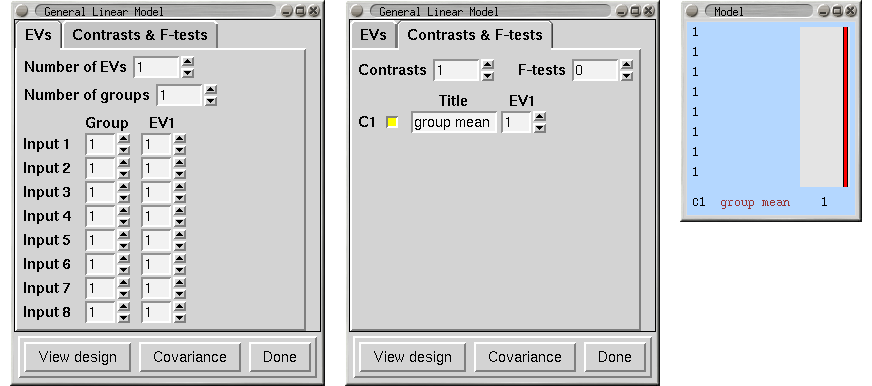
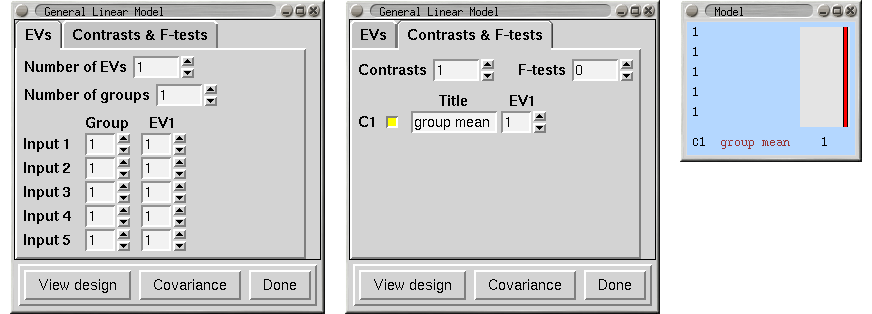
Single-Group Average (One-Sample T-Test)
We have 8 subjects all in one group and want the mean group effect. Does the group activate on average?
|
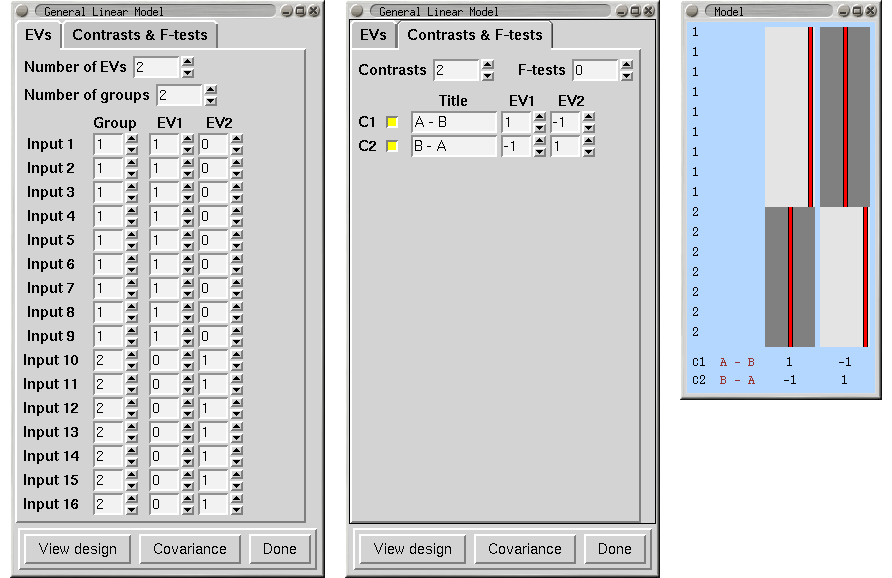
Unpaired Two-Group Difference (Two-Sample Unpaired T-Test)
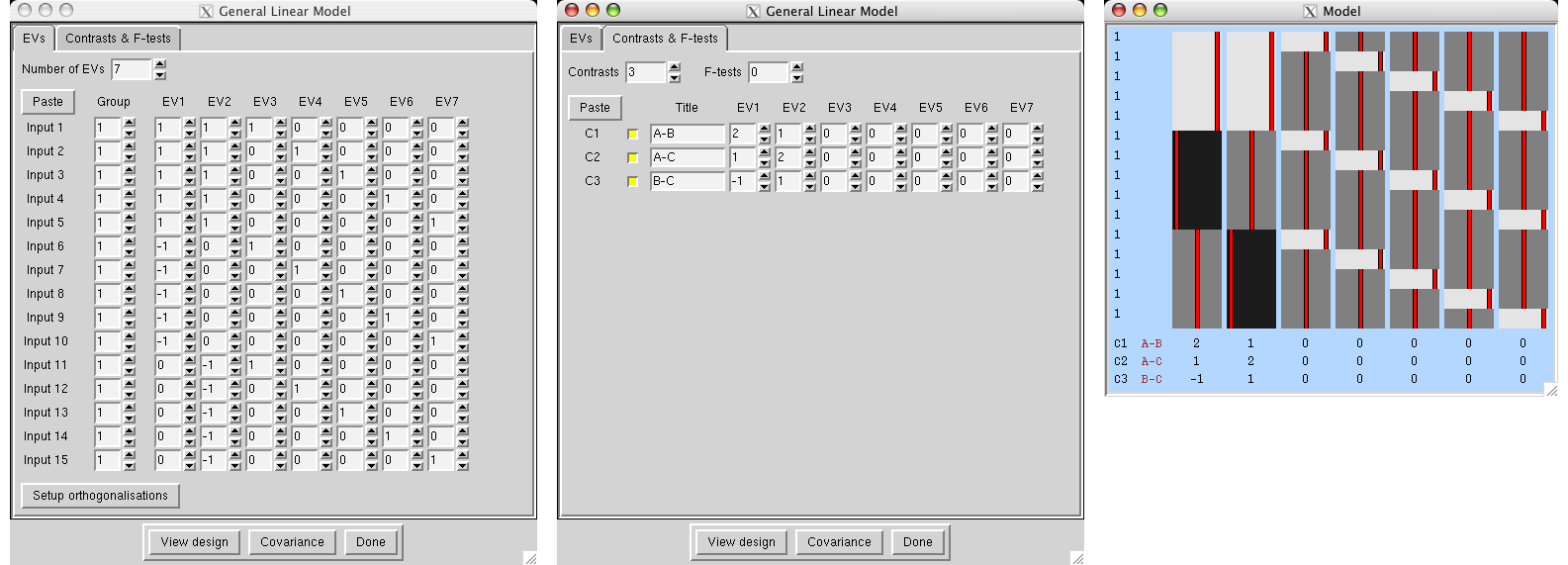
We have two groups of different kinds of subjects (eg 9 patients and 7 controls) with potentially different cross-subject variance in the different groups , so we will specifiy two group "memberships" so that FEAT estimates each group's cross-subject variance separately. We want the mean group difference, and will look in both directions, hence the two contrasts. Note that we cannot setup this model with a single EV (see above).
|
Randomise details
Randomise requires that the grouping variable correspond to "exchangeability blocks" rather than "variance group". Here the randomise example shows all ones for the groupings to indicate the exchangeability rules for the permutations.
|
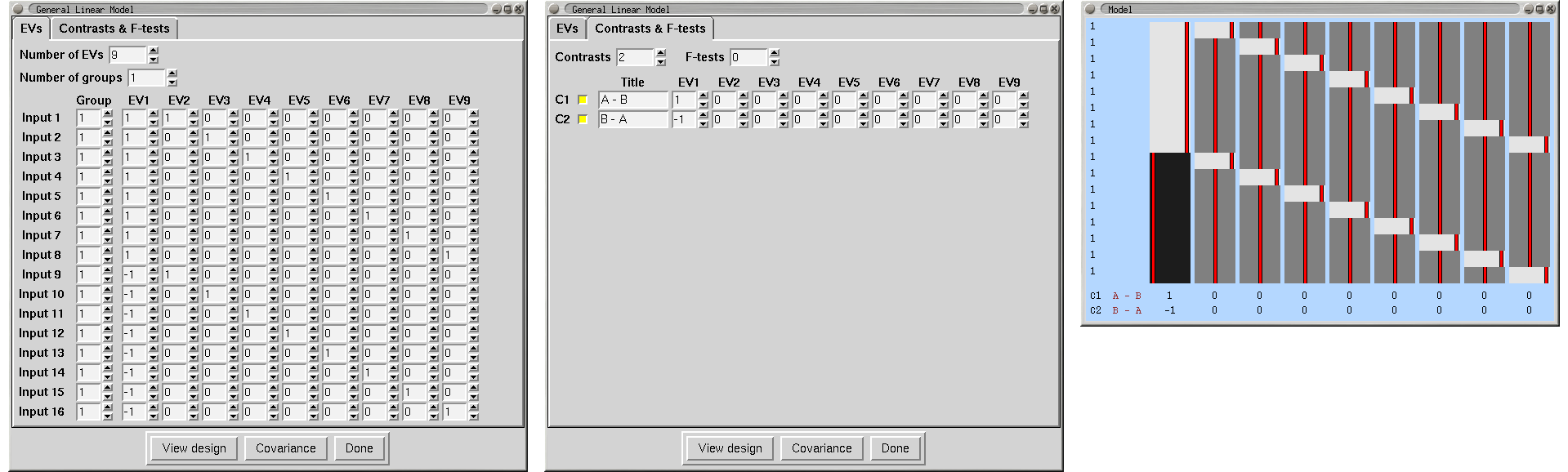
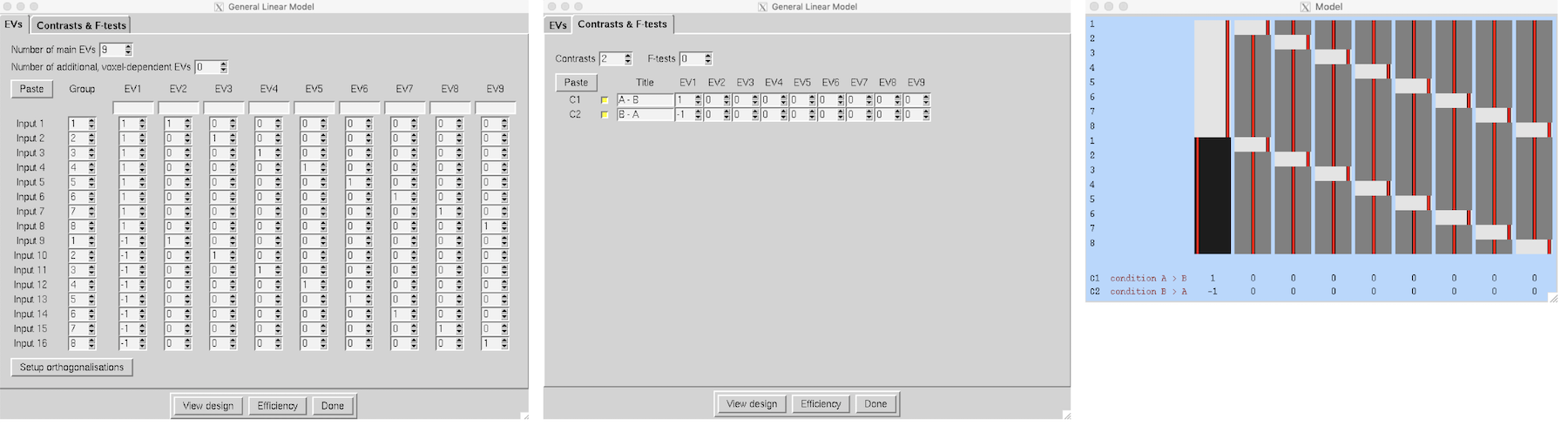
Paired Two-Group Difference (Two-Sample Paired T-Test)
We have a group of 8 subjects scanned under two different conditions, A and B. We enter the condition A analyses as the first 8 inputs, and the condition B analyses as the second 8 inputs. Make sure that the subjects are in the same order within each group of 8! We need one EV for the A-B differences, and then one extra EV for each subject, making 9 in all. EVs 2-9 model each subject's mean effect - in this analysis this is a confound, i.e. parameter estimates 2-9 are ignored, but without this part of the model, the mean effects would intefere with the estimation of the A-B paired differences. A contrast with a one for EV1 and zeros elsewhere tests for A-B paired differences.
|
Randomise details
For the paired t-test, randomise requires a special type of permutation and the most fool-proof way to ensure the test is carried out correctly is to manually compute the differences between runA and runB for each subject and then enter these values into a 1-sample t-test.
As an example, assume that there are 16 separate 3D brain images (for each of the 2 runs for each of the 8 subjects). Start by using fslmaths to compute the difference between runA and runB for each subject and then use fslmerge to create a single 4D volume with 8 images (along the 4th dimension). If working with fMRI, be sure to work with cope and not tstat images.
fslmaths subject1_rA -sub subject1_rB sub1_diff fslmaths subject2_rA -sub subject2_rB sub2_diff (repeat for subjects 3-8) fslmerge -t runA_minus_runB sub1_diff sub2_diff sub3_diff sub4_diff sub5_diff sub6_diff sub7_diff sub8_diff
The resulting 4D volume (i.e. runA_minus_runB) can then be used as the input for the 1-sample t-test, as described above.
When using the GLM GUI or the FEAT GUI, you can also explicitly set the exchangability group values for randomise. If you use this method, you do not need to perform the maths steps above, or setup your analysis as a 1-sample t-test. The screenshot below shows the same design as above, but setup for randomise to exchange subject data correctly.
|
Tripled Two-Group Difference ("Tripled" T-Test)
This is a natural extension of the paired t-test, but the contrasts are slightly counter-intuitive so we explain this case in detail. We have 5 subjects, each scanned under 3 conditions, A, B and C. We enter the 5 condition A scans first, then 5 B and then 5 C. As with the paired t-test, EVs 3-7 simply remove the subject means and are not used in the contrasts.
We now want to form the 3 contrasts A-B, A-C and B-C. Note that, somewhat surprisingly, A-B is not given by [1 0 0...]! We define PE1=a and PE2=b. Then, we can see by looking at the three condition blocks, that the mean (on top of the global mean taken out by EVs 3-7) of condition A is modelled by A=a+b. Likewise, B=-a, and C=-b (look at the values in EVs 1 and 2). Therefore we have A-B = 2a+b = contrast [ 2 1 0....], and A-C = a+2b = contrast [ 1 2 0....], and B-C = -a+b = contrast [ -1 1 0....].
|
Multi-Session & Multi-Subject (Repeated Measures - Three Level Analysis)
5 subjects each have three sessions. For the reasons described above, we will combine across sessions to create COPEs for the subject means of each subject, using a fixed-effects analysis. In the stats GUI, we select Fixed effects. Then we setup the second-level analysis with 5 EVs, where each EV picks out the 3 sessions that correspond to a particular subject. We also need 5 contrasts to represent the 5 subject means, as follows:
|
Now we want the mean group effect, across subjects, achieved with a third-level ME analysis. Select Inputs are lower-level FEAT directories and select the 5 relevant directories created at second-level, named something like subject_N.gfeat/cope1.feat.
|
F-Tests
For example, three groups of subjects, with the question - do the group averages account some significant effect?
|
Single-Group Average with Additional Covariate
We have 7 subjects all in one group. We also have additional measurements such as age, disability scale or behavioural measures such as mean reaction times. The additional effect of the extra measures can be found by entering this as an extra EV, which should be orthogonal wrt the group mean EV - so in this case simply demeaned:
|
ANOVA: 1-factor 4-levels
We have 8 subjects and 1 factor at 4 levels A, B, C and D. The first two inputs are subjects in condition A, the next two are B etc.
To compare a level with another we could just have one EV per level. However, if we want to ask the ANOVA question - where is there any treatment effect then we can do the following. EV1 fits condition D (it is the only nonzero EV during condition D). EV2 fits A relative to this, i.e. represents A-D (see below for explanation). The F-test then tests for any deviation - ie any difference between the levels, and corresponds exactly to the standard ANOVA test.
If, as well as the ANOVA test, you wanted to interpret individual contrasts: If we define m,a,b,c as the 4 PE values, then
A=a+m, B=b+m, C=c+m, D=m.
Thus the first PE, m, is level D, the second is a=A-m=A-D, etc.
To get the mean: mean = (A+B+C+D)/4 = m+(a+b+c)/4 = contrast [ 1 1/4 1/4 1/4 ]
To get A-mean: A-mean = (3a-b-c)/4 = contrast [ 0 3/4 -1/4 -1/4 ] etc.
To get D-mean: D-mean = (-a-b-c)/4 = contrast [ 0 -1/4 -1/4 -1/4 ].
|
ANOVA: 1-factor 4-levels (Repeated Measures)
We have 2 subjects with 1 factor at 4 levels; both subjects had sessions at all 4 levels. The design is the same as above, except that we replace EV1 (the global mean) with a separate mean EV for each subject. This decreases the degrees of freedom in the model, but removes the uninteresting subject-mean variability from the residuals. This approach is analogous to the difference between the unpaired and paired two-group t-test examples shown above. For the other ANOVA examples below, it is similarly straightforward to replace the mean EV with multiple mean EVs in this manner, if you have more than one session per subject.
Subject 1's 4 levels are Inputs 1,3,5,7 and subject 2's 4 levels are Inputs 2,4,6,8.
|
ANOVA: 2-factors 2-levels
Fixed Effects We have 8 subjects and 2 factors, each at 2 levels. To carry out a standard ANOVA we use the following, the three F-tests giving the standard ANOVA results for factor A, factor B and the interaction effect. This assumes that both factors are fixed effects.
|
Random Effects
If both factors are random effects then the F-tests for the effects of the factors are different - the denominator in the F is derived from the interaction effect and not the within-cell errors. In this case, the relevant F images for factors A and B can be formed as Fa=fstat1/fstat3 and Fb=fstat2/fstat3. In order to carry this out, first run FEAT using the above design. Then:
cd <featdir>/stats
fslmaths fstat1 -div fstat3 fstata
fslmaths fstat2 -div fstat3 fstatb
ftoz -zout zfstata fstata 1 1
ftoz -zout zfstatb fstatb 1 1
You could then do thresholding on zfstata and zfstatb with easythresh.
Mixed Effects
If one factor is random and the other is fixed then we want a mixed effects analysis. In this case the fstat which needs the different denominator is the one associated with the fixed factor. For example, if factor A is fixed and factor B is random, then fstat2 already gives you the effect of factor B and for factor A you need to create Fa=fstat1/fstat3 as above.
ANOVA: 3-factors 2-levels
Fixed Effects
We have 16 subjects and 3 factors, each at 2 levels. To carry out a standard ANOVA we use the following, the F-tests giving the standard ANOVA results for the factors and their interactions.
|
Random/Mixed Effects
The following table shows how to test for factor effects with various models:
model |
A |
B |
C |
F(A) |
F(B) |
F(C) |
F(AB) |
F(AC) |
F(BC) |
F(ABC) |
1 |
F |
F |
F |
fstat1 |
fstat2 |
fstat3 |
fstat4 |
fstat5 |
fstat6 |
fstat7 |
2 |
R |
R |
R |
|
|
|
fstat4/fstat7 |
fstat5/fstat7 |
fstat6/fstat7 |
fstat7 |
3 |
F |
R |
R |
|
fstat2/fstat6 |
fstat3/fstat6 |
fstat4/fstat7 |
fstat5/fstat7 |
fstat6 |
fstat7 |
4 |
F |
F |
R |
fstat1/fstat5 |
fstat2/fstat6 |
fstat3 |
fstat4/fstat7 |
fstat5 |
fstat6 |
fstat7 |
Perfusion FMRI Analysis
Introduction
It is straighforward to analyse perfusion FMRI data (often also referred to as ASL or arterial spin labelling) with FEAT. The data needs special treatment because each voxel's timeseries alternates between "tag" and "control" conditions, with control timepoints having higher intensity. If there is BOLD signal present (for example because the echo time is not short) this can be modelled as the average of the tag and control. The perfusion (flow) signal can be modelled as the difference between the tag and control conditions; when there is activation, this difference increases.
There are currently two ways of analysing such data with FEAT - subtraction of tag from control (as part of pre-stats processing), or full perfusion signal modelling, where separate EVs model the BOLD signal, the (constant-height) tag-control difference and the modulation of this by the activation. The choice of which approach to take will depend on the data - subtraction can be better if there is certain structured noise in the data, whilst full modelling can give more accurate modelling, as the data does not have to be resampled, so no information is lost through interpolation.
With either approach, initial evidence suggests that it is worthwhile using highpass filtering (with cutoff set according to the same norms as BOLD FMRI analysis).
Full perfusion signal modelling
If you want to fully model the different aspects of the perfusion data, you will need to setup at least 3 EVs. See the example design matrix: 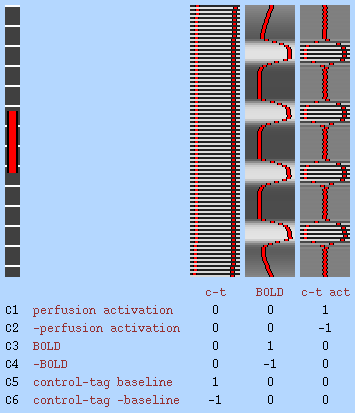
- EV1 models the baseline (constant height) control-tag alternating intensity variation. This models the difference between control and tag conditions when no stimulation is present, i.e., when there is no functional activation.
- EV2 models the BOLD signal, which often contaminates the perfusion signal, particularly if the TE (echo time) is not low. This EV models the slow signal that is the mean of the tag and control conditions. It should be setup as you would setup a normal BOLD FMRI analysis, with the basic EV timings set according to stimulation timings, and then convolved using a standard HRF convolution.
- EV3 is formed by multiplying EVs 1 and 2 together, and models the activation component of the control-tag signal. It is close to zero during rest and rises in amplitude during activation.
For an experiment with just a single experimental condition, these EVs are generally all that you need, and can be setup via the Model setup wizard. If you have more than one condition, then for each new condition you will need a new EV similar to EV2, modelling the BOLD response, and also a corresponding EV formed from interacting the BOLD response EV with EV1. Note that for each interaction EV, you need to set Make zero to Centre for EV1 only (i.e. leave the BOLD EV at Min). This makes the modulation envelope for the perfusion activation EV symmetric about zero, rather than only positive, and results in a slightly better model of the perfusion data.
The above assumes that the tag-control order of the data is such that the first timepoint is tag. If not, you should change the Phase for EV1 from 0 to the TR.
You should leave FILM whitening turned on.
Note that if you use this approach then featquery will not automatically output percent change values that relate to 100*(perfusion signal change)/(perfusion baseline). Instead you need to use featquery to output COPE1 (perfusion activation in the example above) and COPE5 (perfusion baseline in the example above), and then calculate 100*COPE1/COPE5.
Subtraction
Perfusion subtraction subtracts even from odd timepoints in order to convert tag-control alternating timepoints into a perfusion-only signal. If you are setting up a full perfusion model (see above) then you should not use this option. To use the subtraction method, select your input data, and make sure that you set the TR correctly (i.e., how long each tag or control volume takes to acquire) in the Data part of the FEAT GUI. Then, in Pre-stats, setup the pre-processing as normal in FEAT, except that you should turn on Perfusion subtraction. You should not be using intensity normalisation so leave that turned off. You may wish to use Highpass filtering; this will remove low frequency noise still present after the subtraction processing. FEAT will automatically turn off FILM prewhitening if you use perfusion subtraction (this is set in the Stats part of the GUI).
By default it is assumed that your first timepoint is tag, the second is control, etc. If your data is the other way round, change the First timepoint is tag setting to First timepoint is control.
Next, setup the model as you normally would for an FMRI analysis, with an EV for each experimental condition. The subtraction causes a temporal shift of the sampled signal to half a TR earlier; hence you should ideally shift your model forwards in time by half a TR, for example by reducing custom timing onsets by half a TR or by increasing the model shape phase by half a TR.
To see exactly what the subtraction processing is doing, look at the $FSLDIR/bin/perfusion_subtract script. This splits the original timeseries into two, one containing all even timepoints and the other containing all the odds. It then creates two new timeseries from each, in one case shifting the data forwards by half an (original) TR and in the other shifting it back by half a TR. The appropriate combinations of these are then subtracted from each other, before re-combining into a processed dataset that has the same number of timepoints as the original data. Sinc (temporal) interpolation is used in order to minimised temporal blurring induced by the shifting. This approach has better accuracy (with respect to removing BOLD contamination and recovering perfusion signal) and efficiency (particularly with respect to not halving the number of timepoints) than simpler perfusion subtraction methods. However, the filter affects the autocorrelation structure in the timeseries, in a way that is not well-matched to the autocorrelation estimation in FILM. This is why FILM whitening is turned off, and instead, to correct for the reduced degrees of freedom in the filtered data, the varcope and degrees-of-freedom are automatically corrected after running FILM in OLS mode.
Note that if you use this approach then featquery will automatically output percent change values that correspond to 100*(perfusion signal change)/(perfusion baseline).
FEAT Output
FEAT finds the directory name and filename associated with the 4D input data. If the associated directory is writable by you then a related whatever.feat directory is created into which all FEAT output is saved. If not, the FEAT directory is created in your home directory. In either case, if the appropriately named FEAT directory already exists, a "+" is added before the .feat suffix to give a new FEAT directory name. (Of course, all the above gets ignored if you explicitly set the output directory name.)
If you rerun Post-stats or Registration, you can choose (under the Misc tab) whether to overwrite the relevant files in the chosen FEAT directory or whether to make a complete copy of the FEAT directory and write out new results in there.
All results get saved in the FEAT directory.
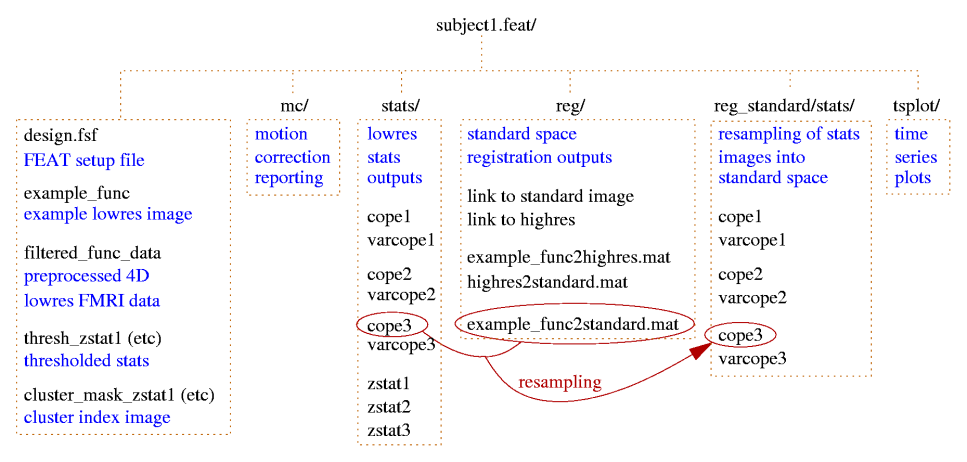
cluster_mask_zstat1 image of clusters found for contrast 1; the values in the clusters are the index numbers as used in the cluster list.
cluster_zstat1.html / .txt the list of significant clusters for contrast 1. Note that the co-ordinates are the original voxel co-ordinates. (MEDx users: MEDx inverts y, so to use the y values in MEDx, use y_medx = y_size - 1 - y_feat where y_size is the size of the image in the y direction.)
cluster_zstat1_std.html / .txt the same, but with co-ordinates given in standard space. This exists if registration to standard space has been carried out.
design.con list of contrasts requested.
design.fsf FEAT setup file, describing everything about the FEAT setup. This can be loaded into the FEAT GUI.
design.fts list of F-tests requested.
design.gif 2D image of the design matrix.
design.mat the actual values in the design matrix.
design.trg event onset times, created to be used in peri-stimulus timing plots.
design_cov.gif 2D image of the covariance matrix of the design matrix.
example_func the example functional image used for colour rendering, and also the one that was used as the target in motion correction. This is saved before any processing is carried out. This is also the image that is used in registration to structural and/or standard images.
filtered_func_data the 4D FMRI data after all filtering has been carried out. (prefiltered_func_data is the output from motion correction and the input to filtering, and will not normally be saved in the FEAT directory.) Although filtered_func_data will normally have been temporally high-pass filtered, it is not zero mean; the mean value for each voxel's timecourse has been added back in for various practical reasons. When FILM begins the linear modelling, it starts by removing this mean.
lmax_zstat1.txt and _std.txt are lists of local maxima within clusters found when thresholding.
mask the binary brain mask used at various stages in the analysis.
rendered_thresh_zstat1.png 2D colour rendered stats overlay picture for contrast 1.
rendered_thresh_zstat1 3D colour rendered stats overlay image for contrast 1. After reloading this image, use the Statistics Colour Rendering GUI to reload the colour look-up-table.
report.log a log of all the programs that the feat script ran (ie the same as report.log but without the log outputs).
report.html the web page FEAT report (see below).
report.log a log of the FEAT run, including all calls to FSL programs and their log outputs.
thresh_zstat1 the thresholded Z statistic image for contrast 1.
mc/prefiltered_func_data_mcf.par a text file containing the rotation and translation motion parameters estimated by MCFLIRT, with one row per volume.
mc/mc_rot.gif, mc/mc_trans.gif plots showing these parameters as a function of volume number (i.e., time).
reg/example_func2highres.* files are related to the registration of the low res FMRI data to the high res image. The .mat file is the transformation in raw text format. The .gif image includes several slices showing overlays of the two images combined after registration. (Note that the inverse of each transform file is also saved (e.g. highres2example_func.mat) to make it easy for you later to take the highres image back into the lowres space.)
reg/example_func2standard.* files are related to the registration of the low res FMRI data to the standard image.
reg/highres is a symbolic link to the high res image.
reg/highres2standard.* files are related to the registration of the high res image to the standard image.
reg/standard is a symbolic link to the standard image.
stats/contrastlogfile a logfile showing how well the statistics fit a Gaussian distribution, on the assumption of no activation.
stats/cope1 the contrast of parameter estimates image for contrast 1.
stats/corrections the normalised covariance of the EV estimates; there is one volume for each element of the nEV x nEV covariance; it is normalised in that the actual covariance requires scaling by the residual variance, sigmasquares.
stats/dof the mean estimated degrees-of-freedom over the whole data set.
stats/neff1 a statistical correction image for contrast 1.
stats/glslogfile a FILM run logfile.
stats/pe1 the parameter estimate image for EV 1.
stats/probs a list of probabilities used for estimating Gaussian statistical fitting.
stats/ratios a list of estimates for Gaussian statistical fitting.
stats/sigmasquareds the 3D image of the residual variance from the linear model fitting.
stats/smoothness the estimation of the smoothness of the 4D residuals field, used in inference.
stats/stats.txt another FILM logfile.
stats/threshac1 The FILM autocorrelation parameters.
stats/tstat1 the T statistic image for contrast 1 (=cope/sqrt(varcope)).
stats/varcope1 the variance (error) image for contrast 1.
stats/zstat1 the Z statistic image for contrast 1
tsplot/tsplot_zstat1.gif the full model vs data plot for the maximum Z statistic voxel from contrast 1.
tsplot/tsplot_zstat1p.gif the plot of reduced data vs cope partial model fit - i.e. data-full_model+partial_model vs partial_model.
tsplot/tsplot_zstat1.txt text file of values used for the above plots, for the maximum Z statistic voxel from contrast 1. The first column is the data, the second column is the partial model fit for contrast 1, the third column is the full model fit and the fourth column is the reduced data for contrast 1.
tsplot/tsplotc_zstat1.* plots and text file as above, but instead of using the peak Z stat voxel, here the data is averaged over all voxels in all significant clusters for contrast 1.
ps_tsplot_zstat1_ev1.gif (etc); peristimulus text files and plots, showing model fits averaged over multiple stimulus repeats and "data scatter" over these repeats. The format for the text files is the same as above, except for the insertion of an extra first column which encodes the peristimulus timing (i.e., time relative to the start time within the peristimulus window). The _ev1 part of the filename means that this file/plot relates to EV1; separate plots and text files are generated for each EV for every contrast.
If you have run F-tests as well as T-tests then there will also be many other files produced by FEAT, with filenames similar to those above, but with zfstat appearing in the filename.
The web page report includes the motion correction plots, the 2D colour rendered stats overlay picture for each contrast, the data vs model plots, registration overlay results and a description of the analysis carried out.
Higher-Level FEAT Output
A second-level .gfeat directory contains one 4D cope* for each of the first-level contrasts; these are simply the concatenation (across the first-level analyses) of those first-level cope images (in standard space) - ie the 4th dimension here is the number of first-level analyses. This is the input to the second-level analysis.
After the second-level analysis has completed each of those 4D cope* files in the .gfeat directory will have resulted in a .feat second-level output, containing all analysis steps (and of course, second-level output copes).
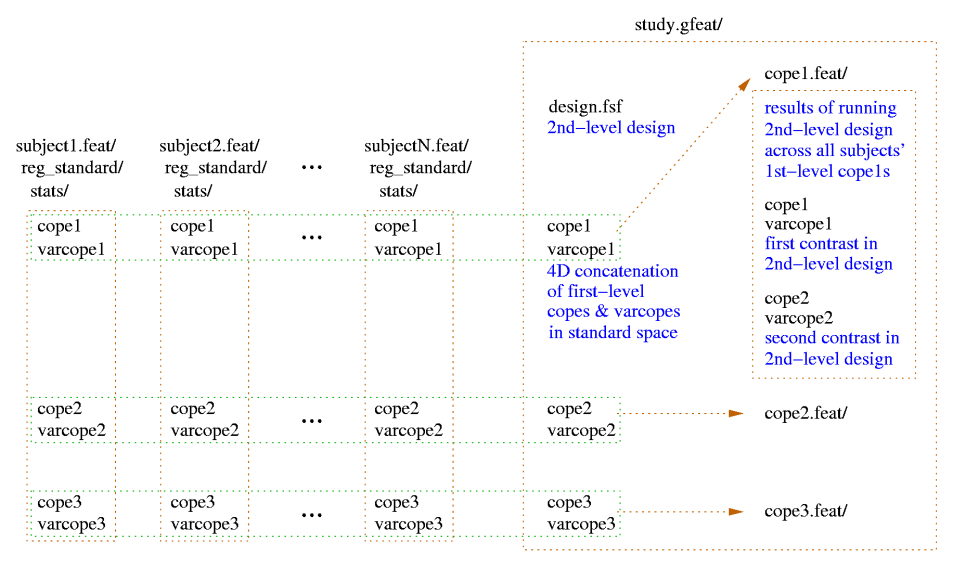
Additional files created include:
cope1.feat/filtered_func_data the 4D file that collects the COPEs from the first level analyses, for contrast 1.
cope1.feat/var_filtered_func_data the 4D file that collects the VARCOPEs from the first level analyses, for contrast 1.
cope1.feat/stats/mean_random_effects_var1 the estimate of the pure between-subjects random effect variance, for contrast 1.
cope1.feat/stats/weights1 4D file that is the reciprocal of the sum of of the 1st level VARCOPES and pure between-subjects random effect variance; the square root of this file are the weights used to whiten data and model as part of Generalised Least Squares, for contrast 1.
FEAT Programs
Feat - the FEAT GUI, as described above.
feat - the FEAT script which the GUI calls when the setup has been completed and GO is pressed. If you have saved a FEAT setup file (eg design.fsf) to file, then you can run FEAT without the GUI by typing feat <featsetupfile.fsf>.
Glm - a GUI for setting up just the design matrix and contrasts, in the same way as in FEAT, for use with other modelling/inference programs such as randomise.
easythresh - a simple script for carrying out cluster-based thresholding and colour activation overlaying.
featregapply - a program which applies the lowres to standard-space registration transforms to the lowres stats images in a FEAT directory to generate standard-space versions, for inputting to higher-level analysis.
Featquery (GUI) and featquery (command-line script) - a program which allows you to interrogate FEAT results by defining a mask or set of co-ordinates (in standard-space, highres-space or loweres-space) and get mean stats values and time-series.
feat_model - a program which uses the FEAT setup file to create the required design matrix as a text file, as well as the related design matrix pictures.
mccutup - convert the estimates of motion saved in a FEAT directory to separate files which can then be fed into FEAT as Custom (1 entry per volume) EVs.
Renderhighres (GUI) and renderhighres (command-line script) - transforms all thresholded stats images in a FEAT directory into high resolution or standard space and overlays these onto the high resolution or standard space images. This then produces PNG format pictures of the overlays and, by default, deletes the 3D AVW colour overlay images.
tsplot - create time series plots for a FEAT directory.
Featquery - FEAT Results Interrogation
Introduction
Featquery is a tool which allows you to interrogate FEAT results within a mask or at a given co-ordinate. The mask/co-ordinate can be in standard-space, highres-space or lowres-space. Within this mask (or at the given co-ordinate), Featquery calculates a number of image and timeseries statistics. For example, you might define a standard-space mask for the motor cortex, and Featquery will tell you the mean (and peak) % signal change associated with your modelled experimental paradigm within that area.
Choosing FEAT directories to interrogate
First you must select a previously-created FEAT output directory whose results you wish to investigate. You can run multiple queries by changing the Number of FEAT directories from the default of 1. The results of running Featquery will be saved in a directory (by default named "featquery") inside each FEAT directory that you select here. If you have already run Featquery on a FEAT directory, a "+" will be appended to the Featquery output name, e.g. "featquery+".
Choosing stats images to investigate
Once you have selected a FEAT directory, a list of stats images inside that FEAT directory will appear. Click the buttons next to those that you are interested in. For each of these in turn, Featquery will calculate various quantities within the selected mask, for example, the number of non-zero voxels within the mask, the mean and max values of the stats image within the mask, and the co-ordinates of the max image value within the mask.
Setting up a mask or voxel co-ordinates
You must now choose a Mask image. This would normally be a binary image in standard-space, highres-space or lowres-space, with a region-of-interest (ROI), for example, the visual cortex, created by any method (e.g. hand-drawn or activation from a multi-subject higher-level FEAT analysis). Featquery will automatically detect which space this mask is in (standard-space, highres-space or lowres-space) and will transform it into the native lowres space of example_func; of course this can only work if FEAT registration was setup and carried out.
If you want a different mask for each selected FEAT directory, specify a mask name as a relative filename (i.e., without a "/"). This mask will then be looked for in each FEAT directory.
Alternatively, you can specify a single co-ordinate (in voxels or mm) at which to extract values from the chosen stats images. This still requires a "mask" image to be chosen, as the co-ordinate is specified in the space of the mask. Thus if you want the co-ordinates to be in lowres space, just select "mask" or "example_func" from the FEAT directory. If you want to specify standard-space co-ordinates (e.g. MNI152 space) then choose "reg/standard". If you choose to specify the co-ordinate in mm (instead of voxels) and the mask image is one with a co-ordinate origin set (e.g. reg/standard which by default is in MNNI152 space, with the origin at the anterior commissure) then the co-ordinates that you enter will be relative to this origin, not the corner of the volume.
Finally, you can also automatically generate a mask from one of the various standard-space atlases provided with FSL. To do this, select an atlas from the menu, and then select a structure by pressing the Select label button.
Featquery saves the mask, once transformed into the native space of the selected FEAT directory, inside the Featquery output directory, called "mask" (even if this only contains a single non-zero voxel, in the case of selecting a co-ordinate).
Further options
If you select an atlas, extra information will be added into the Featquery output. For each stats image, the position of the maximum voxel (within the input mask) will be used to query the selected atlas, and each structure which overlaps this voxel will be reported (for the probabilistic atlases, the % probability for each overlapping structure will be given).
If you select Convert PE/COPE values to % (VARCOPE to %^2), any PE or COPE parameter estimate or contrast values will be converted to percentage signal change values before reporting. This is achieved by scaling the PE or COPE values by (100*) the peak-peak height of the regressor (or effective regressor in the case of COPEs) and then by dividing by mean_func (the mean over time of filtered_func_data). If you are running Featquery on a higher-level FEAT directory, mean_func is the mean of all the lower-level mean_func images, so the % change values are reported correctly. For VARCOPE variance images, the equivalent transformation is also made, this time in units of % squared (i.e., take the square root to compare with the size of the COPE % change).
If you turn on Do not binarise mask (allow weighting) then if your mask is non-binary, its non-zero values will weight all Featquery output values rather than treating the mask as binary. In this case the mask will be scaled such that its mean (over non-zero voxels) is 1.
If you have selected a mask image in standard or highres space, this will get transformed into lowres space as described above. This involves interpolation; at the edges of the mask there will be a continuous range of values from 1 down to 0. In order to get back to a binary mask, this must be thresholded at some value - the default is 0.5. However, if you want the mask to be slightly more or less inclusive than that default, you can Change post-interpolation thresholding of mask - for example, by reducing the value to 0.3, the final lowres mask will be slightly larger.
If, as well as masking the stats images that you selected above, you want to only consider voxels from the images that are above above a given threshold, then turn on Threshold stats images as well as masking and select a value.
By default the output from Featquery is saved in a directory called "featquery", created inside each selected original FEAT directory (or is called "featquery+", etc., if you have already run Featquery before). At Featquery output directory name you can change this default name to anything else in order to more easily distinguish different runs of Featquery.
Go
When you press Go, Featquery produces a web page report with all requested stats, as well as a raw text version of the main report table. Above the main table are links to "Mean time series (masked/weighted)" (this is a raw text file containing the mean time series, within the mask, of the 4D data file filtered_func_data) and "Masked time series plots" (this is a full graphical time series report, similar to those shown in the FEAT report, but with everything masked by the Featquery mask/co-ordinate, including peri-stimulus plots).
Featquery output
The first column in the main table shows the "stats image", i.e. lists the different FEAT stats images that you asked Featquery to report on; each of these is also a link to a raw text file giving the data timeseries plot at the position of the maximum image value within the mask. The next column tells you the number of non-zero voxels within the mask.
The next group of columns give various statistics derived from each image's values within the Featquery mask. Most of these are self-explanatory (min, mean, median, max); the 10% and 90% columns show the image values at 10% and 90% of the way through the ordered list of values (i.e., at 10%/90% of the cumulative distribution function) - so these could be considered a "robust range" of the data values, ignoring the tails/outliers of the distribution.
The final group of columns gives the co-ordinates of the maximum image voxel within the Featquery mask, both in voxels in native space and in mm in standard space (assuming that FEAT registration was run), as well as overlapping atlas strucures, if requested.
Scripting the running of Featquery and the extraction of values generated by Featquery
When you press Go, if you started the Featquery GUI from a command-line terminal then you will see the command-line call to the featquery script that is made. You can easily adapt this to script Featquery, if you want to make lots of calls to featquery automatically. Type "featquery" to get the full command-line usage.
It is easy to extract data from the raw text output, using unix commands like grep and awk. For example, to extract the "# featdir" value (column 1) and the median (column 7) of the "cope2" image, you might use:
cat ~/grot.feat/featquery/report.txt | grep stats/cope2 | awk '{print $1 " " $7}'
Appendix A: Brief Overview of GLM Analysis
General Linear Modelling (more correctly known simply as "linear modelling") sets up a model (i.e., what you expect to see in the data) and fits it to the data. If the model is derived from the stimulation that was applied to the subject in the MRI scanner, then a good fit between the model and the data means that the data was indeed caused by the stimulation.
The GLM used here is univariate. This means that the model is fit to each voxel's time-course separately. (Multivariate would mean that a much more complex analysis would take place on all voxels' time-courses at the same time, and interactions between voxels would be taken into account. Independent Component Analysis - ICA - is an example of multivariate analysis.) For the rest of this section, you can imagine that we are only talking about one voxel, and the fitting of the model to that voxel's timecourse. Thus the data comprises a single 1D vector of intensity values.
A very simple example of linear modelling is y(t)=a*x(t)+b+e(t). y(t) is the data, and is a 1D vector of intensity values - one for each time point, i.e., is a function of time. x(t) is the model, and is also a 1D vector with one value for each time point. In the case of a square-wave block design, x(t) might be a series of 1s and 0s - for example, 0 0 0 0 0 1 1 1 1 1 0 0 0 0 0 etc. a is the parameter estimate for x(t), i.e., the value that the square wave (of height 1) must be multiplied by to fit the square wave component in the data. b is a constant, and in this example, would correspond to the baseline (rest) intensity value in the data. e is the error in the model fitting.
If there are two types of stimulus, the model would be y=a1*x1+a2*x2+b+e. Thus there are now two different model waveforms, corresponding to the two stimulus timecourses. There are also two interesting parameters to estimate, a1 and a2. Thus if a particular voxel reponds strongly to model x1 the model-fitting will find a large value for a1; if the data instead looks more like the second model timecourse, x2, then the model-fitting will give a2 a large value.
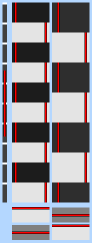 GLM is normally formulated in matrix notation. Thus all of the parameters are grouped together into a vector A, and all of the model timecourses are grouped together into a matrix X. However, it isn't too important to follow the matrix notation, except to understand the layout of the design matrix, which is the matrix X. The main part of the image shown here contains two such model timecourses. Each column is a different model timecourse, with time going down the image vertically. Thus the left column is x1, for example, the timecourse associated with visual stimulation, and the right column is x2, e.g., auditory stimulation, which has a different timecourse to the visual stimulation. Note that each column has two representations of the model's value - the black->white intensity shows the value, as does the red line plot. Make sure that you are comfortable with both representations.
GLM is normally formulated in matrix notation. Thus all of the parameters are grouped together into a vector A, and all of the model timecourses are grouped together into a matrix X. However, it isn't too important to follow the matrix notation, except to understand the layout of the design matrix, which is the matrix X. The main part of the image shown here contains two such model timecourses. Each column is a different model timecourse, with time going down the image vertically. Thus the left column is x1, for example, the timecourse associated with visual stimulation, and the right column is x2, e.g., auditory stimulation, which has a different timecourse to the visual stimulation. Note that each column has two representations of the model's value - the black->white intensity shows the value, as does the red line plot. Make sure that you are comfortable with both representations.
When the model is fit to the data, for each voxel there will be found an estimate of the "goodness of fit" of each column in the model, to that voxel's timecourse. In the visual cortex, the first column will generate a high first parameter estimate, and the second column will generate a low second parameter estimate, as this part of the model will not fit the voxel's timecourse well. Each column will be known as an explanatory variable (EV), and in general represents a different stimulus type.
To convert estimates of parameter estimates (PEs) into statistical maps, it is necessary to divide the actual PE value by the error in the estimate of this PE value. This results in a t value. If the PE is low relative to its estimated error, the fit is not significant. Thus t is a good measure of whether we can believe the estimate of the PE value. All of this is done separately for each voxel. To convert a t value into a P (probability) or Z statistic requires standard statistical transformations; however, t, P and Z all contain the same information - they tell you how significantly the data is related to a particular EV (part of the model). Z is a "Gaussianised t", which means that a Z statistic of 2 is 2 standard deviations away from zero.
As well as producing images of Z values which tell you how strongly each voxel is related to each EV (one image per EV), you can compare parameter estimates to see if one EV is more "relevant" to the data than another. This is known as contrasting EVs, or producing contrasts. To do this, one PE is subtracted from another, a combined standard error is calculated, and a new Z image is created. All of the above is controlled by you, by setting up contrasts. Each output Z statistic image is generated by setting up a contrast vector; thus set the number of outputs that you want. To convert a single EV into a Z statistic image, set it's contrast value to 1 and all others to 0. Thus the simplest design, with one EV only, has just one contrast vector, and only one entry in this contrast vector: 1. To compare two EVs, for example, to subtract one stimulus type (EV 1) from another type (EV 2), set EV 1's contrast value to -1 and EV 2's to 1. A Z statistic image will be generated according to this request, answering the question "where is the response to stimulus 2 significantly greater than the response to stimulus 1?"
The bottom part of the above image shows the requested contrasts; each column refers to the weighting of the relevant EV (often either just 1, 0 or -1), and each row is a different contrast vector. Thus each row will result in it's own Z statistic image. Here the contrasts are [1 0] and [0 1]. Thus the first Z stat image produced will show response to stimulus type 1, relative to rest, and the second will show the response to stimulus type 2.
 If you want to model nonlinear interactions between two EVs (for example, when you expect the response to two different stimuli when applied simultaneously to give a greater response than predicted by adding up the responses to the stimuli when applied separately), then an extra EV is necessary. The simplest way of doing this is to setup the two originals EVs, and then add an interaction term, which will only be "up" when both of the original EVs are "up", and "down" otherwise. In the example image, EV 1 could represent the application of drug, and EV 2 could represent visual stimulation. EV 3 will model the extent to which drug+visual is greater than the sum of drug-only and visual-only. The third contrast will show this measure, whilst the fourth contrast [0 0 -1] shows where negative interaction is occurring.
If you want to model nonlinear interactions between two EVs (for example, when you expect the response to two different stimuli when applied simultaneously to give a greater response than predicted by adding up the responses to the stimuli when applied separately), then an extra EV is necessary. The simplest way of doing this is to setup the two originals EVs, and then add an interaction term, which will only be "up" when both of the original EVs are "up", and "down" otherwise. In the example image, EV 1 could represent the application of drug, and EV 2 could represent visual stimulation. EV 3 will model the extent to which drug+visual is greater than the sum of drug-only and visual-only. The third contrast will show this measure, whilst the fourth contrast [0 0 -1] shows where negative interaction is occurring.
All of the EVs have to be independent of each other. This means that no EV can be a sum (or weighted sum) of other EVs in the design. The reason for this is that the maths which are used to fit the model to the data does not work properly unless the design matrix is of "full rank", i.e. all EVs are independent. A common mistake is to model both rest and activation waveforms, making one an upside-down version of the other; in this case EV 2 is -1 times EV 1, and therefore linearly dependent on it. It is only necessary to model the activation waveform.
With "parametric designs", there might be several different levels of stimulation, and you probably want to find the response to each level separately. Thus you should use a separate EV for each stimulation level. (If, however, you are very confident that you know the form of the response, and are not interested in confirming this, then you can create a custom waveform which will match the different stimulation levels, and only use one EV.) If you want to create different contrasts to ask different questions about these responses, then: [1 0 0] shows the response of level 1 versus rest (likewise [0 1 0] for level 2 vs rest and [0 0 1] for level 3). [-1 1 0] shows where the response to level 2 is greater than that for level 1. [-1 0 1] shows the general linear increase across all three levels. [1 -2 1] shows where the increase across all three levels deviates from being linear (this is derived from (l3-l2)-(l2-l1)=l3-2*l2+l1).
Thus there often exists a natural hierarchy in contrast vectors. In the above example, [1 1 1] shows overall activation, [-1 0 1] shows linear increase in activation and [1 -2 1] shows (quadratic) deviation from the linear trend. Note that each contrast is orthogonal to the others (e.g. -1*1 + 0*1 + 1*1 = 0) - this is important as it means that each is independent of the others. A common mistake might be, for example, to model the linear trend with [1 2 3], which is wrong as it mixes the average activation with the linear increase.
Appendix B: Design Matrix Rules
This section describes the rules which are followed in order to take the FEAT setup and produce a design matrix, for use in the FILM GLM processing.
Here HTR model means "high temporal resolution" model - a time series of values that is used temporarily to create a model and apply the relevant HRF convolution before resampling down in time to match the temporal sampling of the FMRI data.
Note that it is assumed that every voxel was captured instantaneously in time, and at the same time, exactly halfway through a volume's time period, not at the beginning. This minimises timing errors, if slice-timing correction has not been applied.
No constant column is added to the model - instead, each EV is demeaned, and each voxel's time-course is demeaned before the GLM is applied.
for each EV
(
if ( square waveform )
fill HTR model with 0s or 1s
else if ( sinusoidal waveform )
fill HTR model with sinusoid scaled to lie in the range 0:1
else if ( custom waveform )
fill HTR model with custom information, with 0s outside of
specified periods
demean
create "triggers" i.e. record the start and end of event or block
create blurring+delaying HTR HRF convolution kernel, normalised so
that the sum of values is 1 (in the case of basis functions, several
related kernels are created)
convolve HTR model with HRF convolution kernel (values in HTR model
for t<0 are set to 0 to allow simple convolution)
OR
in the case of sinusoidal original waveform; create harmonics (if
requested)
subsample HTR model to match the temporal resolution of the data;
take the value in the centre of each volume's time period
add motion parameters as confound EVs if requested on the command
line
do two passes
(
take model produced by subsampling step above
if pass=2 then apply high-pass temporal filtering
re-demean
instead of all the above - if this EV is an "interaction"
(nonlinear interaction between other EVs);
model = PRODUCT(other EVs, after subtracting the min value from
each)
orthogonalise current EV wrt earlier EVs if requested (form
temporary matrix from selected EVs, carry out SVD, and subtract
projection of current EV onto each vector in SVD output)
orthogonalise main model wrt motion parameter EVs if requested
if requested, create a new EV, calculated as the temporal derivative
of the current EV
re-demean
if pass=1 then estimate peak-peak model heights
if pass=2 then estimate contrast estimability information
)
)