Contents
- Introduction
- User Guide
Introduction to TIRL scripts
TIRL is a general-purpose Python library that was designed to aid the prototyping of bespoke image registration pipelines. A pipeline that uses the components of TIRL to perform a specific registration task is hereafter designated as tirlscript. The FSL distribution of TIRL supplies with preinstalled tirlscripts for histology-to-MRI registration that can be executed from the command line, using the fslpython environment.
Tirlscripts are python source files that are located in the scripts package of the installed TIRL library. When creating new pipelines, users are encouraged to take the preinstalled scripts as templates, modify them, and install the new tirlscript under a different name. This allows sharing tirlscripts between fellow TIRL users without overwriting library defaults.
Using the command line interface
Activating the fslpython environment
Main menu
1 tirl
2
3 Tensor Image Registration Library
4 Copyright (C) 2018-2020 University of Oxford
5 Part of the FMRIB Software Library (FSL)
6 Author: Istvan N. Huszar
7
8 Available options:
9 home displays the installation directory
10 version displays the library version
11 config [text editor] modify library-wide constants
12 info <file> display information about a TIRL file
13 module list list available tirl scripts
14 add <module.py> [package] add custom module to the library
15 edit <module.path> edit any module in the library
16 remove <custom.pkg.module> remove a user module from the library
17 <script> [arguments] runs an installed TIRL script
18
tirl home
Displays the path to the TIRL root directory. It should be the same as the path that where the $TIRLHOME environmental variable is pointing. The only exception from this is when you have downloaded the TIRL source files, and you are in that source directory, which is different from the directory where TIRL was installed by FSL. In that case, tirl home will return the current directory where the source files are, and $TIRLHOME will keep pointing to the FSL-installed TIRL directory. If you are working on the source files separately, it is recommended that you create a new conda environment that is separate from fslpython.
tirl version
Displays the version of TIRL. Versioning uses the major.minor.revision format. Tirlscripts that were created using a certain major version are not expected to work with a different major version of the library. Minor versions indicate significant changes but maintained compatibility of running previous scripts. Revisions indicate bugfixes that may be important for specific applications but not significant enough for another release.
tirl config
Use this command to modify library-wide constants. Making changes in the library configurations will alter the behaviour of the library. A detailed description of each parameter can be found here. You may leave most things as default, but you must set the temporary working directory (TWD) after installation. The TWD is that TIRL will use as a temporary workspace, where it will deposit large files etc. It is recommended to have this directory on a fast-access storage device, such as an NVME SSD.
tirl info
Displays the header of a file that was generated by TIRL. TIRL files contain a human-readable ASCII header that defines the structure of the object, and a binary part appended after the header, which is non-human readable and contains all relevent numerical data of the object. By inspecting the header, you can see for instance all transformations that were attached to an image that was exported at an intermediate stage of a registration, but you cannot see the parameter values of these transformations. The header can be rather voluminous, so piping to the more shell command is encouraged: tirl info file.timg | more.
tirl module list
Use this command to list all available tirlscripts that can be run from the command line as tirl <scriptname>. Those scripts will be listed here that are in the tirl.scripts or tirl.usermodules package, have a .py extension, and contain the following variable definition: __tirlscript__ = True.
tirl module add
Use this command to install a new tirlscript or if you would like to add a new dependency module to the TIRL installation. Typically, you will need to install a new tirlscript, if you created a new pipeline for a specific application, or made modifications to an existing one that you would like to save under a new name. A typical example of installing a new dependency module would be providing custom image processing steps prior to registration. For example, the configuration file of the slice_to_volume tirlscript defines preprocessing steps for each input image, which are performed before the registration begins. You may specify a custom preprocessing step as myoperations.myfilter, where myoperations is a dependency module that you installed using this command, and myfilter is a function that is defined in myoperations. A new module will be installed in the usermodules package by default, but you can install it to scripts by typing tirl module add myscript scripts. For new or modified tirlscripts, please remember that you have to designate your source file as tirlscript by including the line __tirlscript__ = True.
tirl module edit
Allows quick editing access to installed tirlscripts and dependency modules. The target module must be specified by its absolute path within TIRL, e.g. scripts.mnd.slice_to_volume. This will open the requested python source file in vim. To use a different text editor, append the name of its executable to the end of the command, e.g. for Atom, type tirl module edit scripts.mnd.slice_to_volume atom. Files that are edited this way do not have to be reinstalled to TIRL - the changes will take effect as soon as you save the file. This editing functionality is intended for quick changes such as bugfixes. For more elaborate tasks, such as writing a module from scratch, it is recommended that you load the TIRL installation with a suitable IDE, such as pycharm, and edit files from there.
tirl module remove
Removes a tirlscript or a dependency module from the TIRL installation. The target module must be specified by its absolute path within TIRL, such as scripts.mnd.slice_to_volume. No backups are kept within the library, so make sure only remove a dependency module or tirlscript from TIRL if you are confident that it is no longer needed.
Running tirlscripts
To run a tirlscript from the command line, the tirlscript must have been previously installed into TIRL. The only exceptions from this are the preinstalled scripts, which you can find by typing tirl module list. To execute a script, type tirl <scriptname> [args] in the command line, where <scriptname> is the name of the tirlscript without the .py extension, and [args] are any optionally specified arguments for the tirlscript. If you execute preinstalled tirlscripts without arguments, a help message will be displayed showing the usage of the script, including any arguments that you may specify.
Histology-to-MRI registration
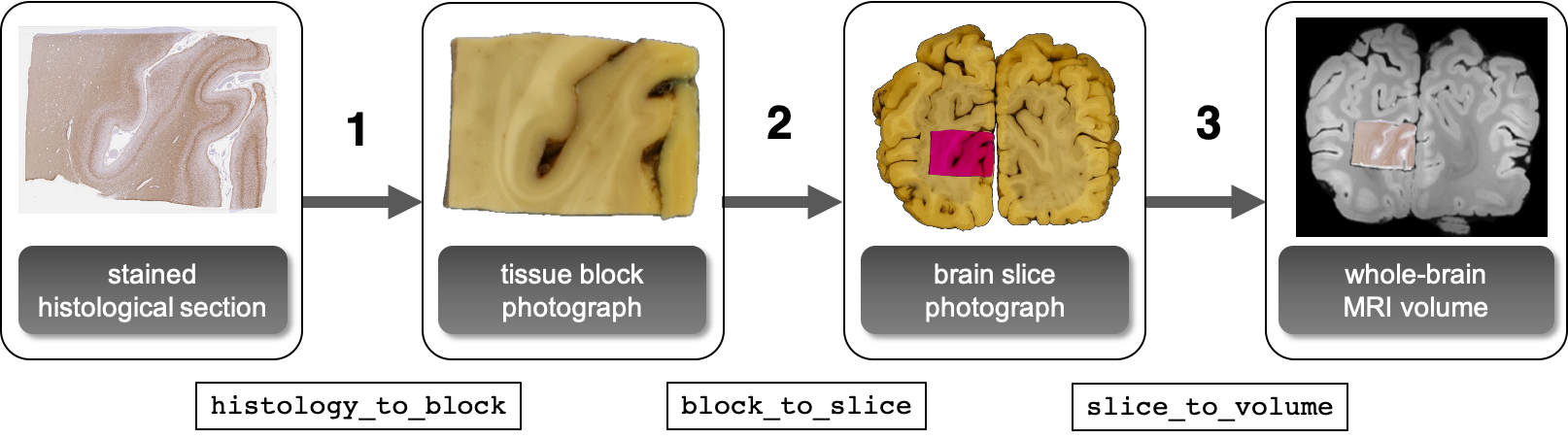
Histology-to-MRI registration is implemented in three stages, with the help of photographic intermediates. Intermediates are useful for the automation of the registration process. If not available, they can be replaced by suitable manual initialisation of the relevant parameters, using only the stage-1 tirlscirpt (for a 2D-to-2D registration) or the stage-3 tirlscirpt (for a 2D-to-3D registration). The tissue block photograph provides a semi-rigid reference for the histological sections that will shrink as they undergo paraffin embedding, and will deform as the tissue sections are mounted on glass slides. The brain slice photograph intermediate is intended to aid the initialisation of the histological section in MRI space, as the cross-sectional anatomy of the whole brain provides enough information to find the position and orientation of the slice in the volume automatically. End-to-end mapping of histology voxel coordinates to MRI voxel coordinates is realised as a combination of the above three stages. The tirlscripts related to the individual stages as well as the combination script are located in tirl.scripts.mnd.
Stage 1: Histology to tissue block photograph
Registers the image of a stained histological section to the photograph of the tissue block from which the section was taken from.
Input
Running stage 1 requires:
histology image |
This may be in any 2D image format, including the Aperio SVS format, as exported by the slide scanner. |
tissue block photograph |
This may be in any 2D image format. The anatomical pattern of the tissue block must match that of the histology, although the effect of slicing the block at a depth, where the anatomy might be slightly different is a known problem, and it can be compensated using the optional stage 4 tirlscript. See also the section on the slicing depth problem. |
a matching 2D MRI slice |
If not using photographic intermediates, the stage-1 tirlscript can be used to align histology directly with MRI, provided that a suitably chosen 2D slice is extracted from the MRI data and specified in lieu of a tissue block photograph. The anatomical pattern of the MRI slice must match that of the histology. |
foreground mask for the tissue block |
A binary image with 1s over the area of the tissue block, 0s elsewhere. If the tissue block image is segmented (backgorund is 0), the mask can be generated automatically by the script based on intensity thresholding. To generate background-segmented images, the background should have a distinct colour from the brain tissue. Please have a look at the background segmentation script supplied with TIRL: stripbg. |
output directory |
Full path to the output directory. It will be created if it does not yet exist. It is recommended to keep this separate from other files, and use the directory merely to contain the result of stage 1 to avoid any interference between different tirlscripts. |
configuration file |
Definition of parameters for the substages of the stage-1 registration. See a detailed description of the parameters here. |
All of the above inputs can be specified within the configuration file, in which case it is sufficient to run this script by specifying the path to the customised configuration file:
A template configuration file is provided in $TIRLHOME/scripts/mnd/cnf_histology_to_block.json. For many applications you may find that the default values will provide a sufficiently good alignment. If you need to change some of the parameters to improve the result of the registration, you should take a look at the Further Information page of TIRL to familiarise yourself with the substages of the histology-to-block registration, and also at this page that gives a detailed description of the each parameter.
Orientation warning: Users should actively pay attention to the orientation of their 2D and 3D images. Capturing a photograph of the posterior surface of a coronal brain slice yields NEUROLOGICAL image orientation, whereby the left hand side of the image represents the left hemisphere and vice versa. (Provided that the image is upright and the horizontal edge of the image aligns with the transverse axis of the brain.). Capturing the anterior view in the same fashion yields an image that is in RADIOLOGICAL orientation, i.e. the left hand side of the image corresponds to the right hemisphere. To avoid confusion between left and right, it is recommended that all images that are used in the histology-MRI registration pipeline have the same orientation, including the MRI (see also under stage 3). In our experiments we used the NEUOROLOGICAL convention.
Output
configuration.json |
copy of the configuration file that was used at the execution of histology_to_block |
fixed1_rotation.timg |
fixed image (histology) after the initial rotational alignment - saved as a TImage object |
fixed2_rigid.timg |
fixed image (histology) after the rigid-body alignment - saved as a TImage object |
fixed3_affine.timg |
fixed image (histology) after the affine alignment - saved as a TImage object |
fixed4_nonlinear.timg |
fixed image (histology) after the non-linear alignment - saved as a TImage object |
fixed.png |
fixed image (histology) after the preprocessing (before registration) - saved as a snapshot |
fixed.timg |
fixed image (histology) after the preprocessing (before registration) - saved as a TImage object |
histology_lowres.png |
|
histology.timg |
|
logfile.log |
log file (unless you have specified a different name or path for the log file) |
moving1_rotation.png |
moving image (tissue block) after the initial rotational alignment - saved as a snapshot |
moving2_rigid.png |
moving image (tissue block) after the rigid-body alignment - saved as a snapshot |
moving3_affine.png |
moving image (tissue block) after the affine alignment - saved as a snapshot |
moving4_nonlinear.png |
moving image (tissue block) after the non-linear alignment - saved as a snapshot |
moving.png |
moving image (tissue block) after the preprocessing (before registration) - saved as a snapshot |
moving.timg |
moving image (tissue block) after the preprocessing (before registration) - saved as a TImage object |
Stage 2: Tissue block photograph to brain slice photograph
Registers a photograph of a tissue block to a photograph of the corresponding brain slice.
Input
The following inputs are required to run stage 2:
tissue block photograph |
|
brain slice photograph |
|
insertion site coordinates --init y x |
Point in the brain slice photograph where the centre of the tissue block image should be inserted. The voxel coordinates are in (row, column) order, where (0, 0) is the top left corner of the image. Coordinates might be specified directly by manual input on the command line (--init y x). Alternatively, a list of possible insertion points can be specified by a text file (--init site.txt), the lines of which correspond to the different insertion sites. For an automated determination of the insertion coordinates, see the description of find_sites below. |
output directory |
